Phytoplankton are major food sources in aquatic ecosystems and play an important role in the global cycle of carbon and silicon, which has led many researchers to focus their efforts on investigating their diverse characteristics [1]. To research this microscopic organism in detail, an appropriate imaging system is essential. Several optical microscopic techniques have been used for the study of phytoplankton [1, 2]. However, conventional light microscopy techniques have severe drawbacks in imaging and quantitative studying of phytoplankton. For example, bright-field microscopy only produces low imaging contrasts for transparent objects such as phytoplankton. Phase contrast [3] or differential interference contrast (DIC) microscopy [4] provide enhanced imaging contrast for phytoplankton via interference; however, neither the phase contrast nor DIC microscopy provides quantitative information, and they are not capable of 3-D imaging. Although confocal laser scanning microscopy [5] provides the 3-D structures of phytoplankton, samples usually need to be labelled with exogenous agents such as fluorescent proteins or dyes, which cause phototoxicity, photobleaching, and interference with intrinsic cellular metabolism [6].
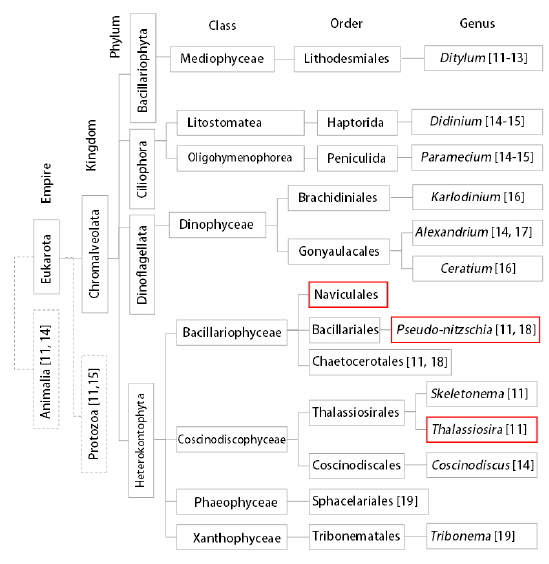
Recently, there has been an emergence of quantitative phase imaging (QPI) or digital holography techniques and their applications in the study of transparent phase objects such as biological cells and tissues [7-13]. The QPI techniques − based on light interferometry − quantitatively measure optical path length shifts introduced by the refractive index (RI) distribution in a specimen. Several QPI techniques have been extensively employed for investigating phytoplankton research, as summarized in Fig. 1. Using digital in-line holographic microscopy or digital holographic microscopy based on Mach-Zehnder interferometry, the shapes, sizes, and 3-D positions of living organelles were measured [14-22]. However, previous works have only focused on the 2-D imaging of phytoplankton and 3-D internal structures of individual live phytoplankton have not been measured yet, largely owing to the limitation of tomographic capability of conventional in-line holography or digital holographic microscopy. We note a notable exception in which 3-D RI maps of non-living diatom skeletons have been measured [23].
Here we present the measurements of high-resolution 3-D RI maps of phytoplankton employing optical diffraction tomography [24-26]. Multiple 2-D QPI images of individual phytoplankton are recorded with various angles of illumination, from which a 3-D RI map of the sample is reconstructed taking into account the effects of optical diffraction, analogous to computed tomography in X-ray. This measurement of a 3-D RI map provides the quantitative structural and biochemical information of phytoplankton including the dry mass, the volume, and the cytoplasmic density of phytoplankton. Furthermore, the measured set of multiple 2-D QPI images of phytoplankton recorded with various angles of illumination was also utilized for generating a high-resolution 2-D QPI image employing the synthetic aperture algorithm [27], which provides the spatial resolution beyond the diffraction limit determined by the numerical aperture (NA) of a used imaging system. The measured 2-D and 3-D quantitative phase images of phytoplankton and the parameters obtained clearly demonstrate the viability of the present method as a useful tool with unique advantages for the study of phytoplankton.
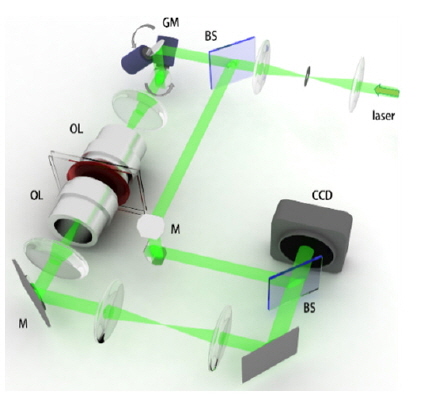
To measure the quantitative phase images of individual phytoplankton, we employed quantitative phase microscopy based on Mach-Zehnder interferometry (Fig. 2). The experimental setup is equipped with a two-axis galvanometer mirror (GM, GVS012/M, Thorlabs, USA), which varies the illumination angle of an incident beam for the tomographic measurement. This setup is conceptually comparable with setups used in previous works [24, 28].
For an illumination source, a diode-pumped solid state (DPSS) laser (
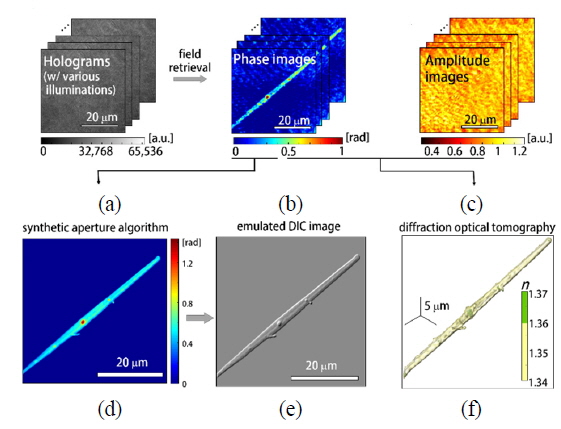
At the microscopic stage, a sample is illuminated by a plane laser beam, and the angle of illumination can be systematically controlled by rotating the two-axis galvanomirror. For one sample, a set of 300 interferograms with various angles of illumination is recorded at a frame rate of 100 Hz. From the set of measured interferograms (Fig. 3(a)), the corresponding phase and amplitude information (Fig. 3(b) and (c)) of the sample can be retrieved using an appropriate phase retrieval algorithm [29]. These holograms measured with different illumination angles are then used to reconstruct the high-resolution 2-D quantitative phase map using synthetic aperture algorithm [27] and 3-D RI tomogram image of a sample employing optical diffraction tomography [24-26], which is explained in detail in the following section.
2.2. Image Reconstruction Processes
In order to construct 2-D high-resolution phase images of phytoplankton (Fig. 3(d), we employed the synthetic aperture imaging algorithm [27]. The principle of the synthetic aperture algorithm is to numerically extend the aperture of an imaging system in order to achieve high spatial resolution and high signal-to-noise ratio (SNR) [30, 31]. Measured holograms of a sample obtained with various illumination angles contain information on the scattering potential of a sample, which cannot be accessed by a conventional optical imaging system with a fixed aperture. Employing the synthetic aperture algorithm, this information can be fully harnessed and reconstructed in the 2-D Fourier space, and finally provides a high-resolution 2-D phase image of a sample beyond the diffraction-limit determined by NA of the optical imaging system used. The representative phase image obtained with the aperture synthesis algorithm is shown in Fig. 3(d).
Compared to the phase image directly measured from the QPI system (Fig. 3(b), the phase image obtained with the aperture synthesis algorithm (Fig. 3(d)) provides higher spatial resolution and SNR ratio. In conventional quantitative phase images, the maximum spatial frequency
In order to construct 3-D RI maps of phytoplankton, we employed an optical diffraction tomography algorithm. Based on the Fourier diffraction theorem [33], a set of measured 2-D optical fields can be mapped onto a 3-D scattering potential spectrum. The spatial frequency information of a 2-D optical field measured with a plane wave with a specific angle of illumination corresponds to a hemispheric area, so called
3.1. Quantitative Holographic Images of Individual Phytoplankton
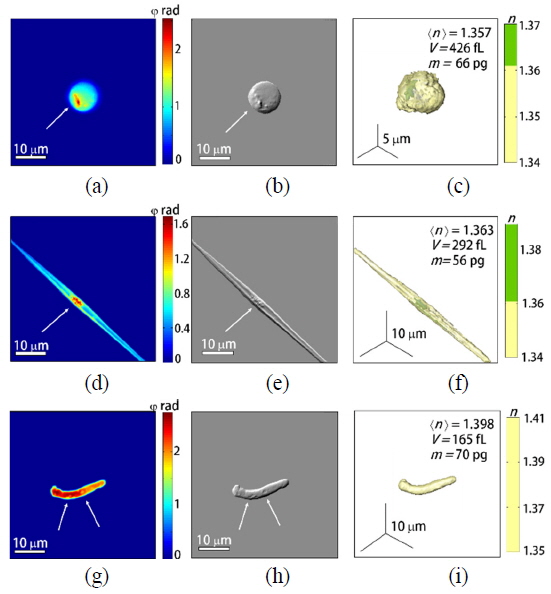
To demonstrate the capability of quantitative imaging of phytoplankton, we measured 4, 11, 12 of spherical-, rod-shaped-, and non-spherical phytoplankton, respectively. These phytoplankton samples were collected from a pond in Daejeon, South Korea. The 50 μL of pond water was directly loaded between two coverslips (24 × 50 mm, Matsunami, Japan) without adding any exogenous agents. The holographic images of representative individual phytoplankton are depicted in Fig. 3. The high-resolution 2-D phase images obtained with the synthetic aperture algorithm (Figs. 4(a), (d), and (g)], the emulated DIC (Figs. 4(b), (e), and (h)) and the 3-D RI distributions (Figs. 4(c), (f), and (i)) of individual spherical-, rod-shaped-, and non-spherical phytoplankton were reconstructed from sets of measured 2-D phase images, respectively (See Methods).
Based on the measured morphology and internal structures, the measured spherical phytoplankton (Fig. 4(a)-(c)) seem to belong to
3.2. Quantitative Phytoplankton Parameters: Volume, Dry Mass, and Density
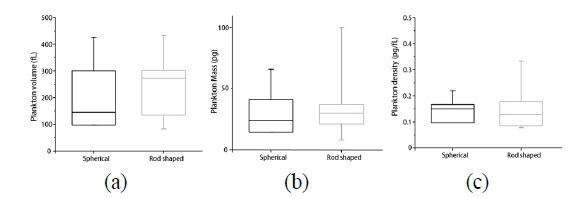
In order to demonstrate quantitative analysis capabilities of quantitative phase imaging of phytoplankton, we retrieved morphological and biochemical properties of individual phytoplankton. The reconstructed 3-D RI maps and 2-D quantitative phase images using the synthetic aperture algorithm allow the quantitative analysis of phytoplankton, which provides various parameters including the volume, dry mass, and cellular density of individual phytoplankton, simultaneously. From the measured volume and dry mass, we also can calculate the density of each phytoplankton. These analyses were performed to 4 spherical and 11 rod-shaped phytoplankton and the results are represented in Fig. 5. We did not conduct statistical analysis for the non-spherical phytoplankton due to their large diversity of sizes and shapes.
The volumes of individual phytoplankton are obtained from the measured 3-D RI maps. The accumulated number of voxels in a 3-D RI map of individual phytoplankton having the RI value greater than that of surrounding media was translated to the total volume considering the volume of a unit voxel. Figure 5(a) presents the volume distribution of the spherical and rod-shaped phytoplankton group. The mean values of cellular volumes of individual phytoplankton are 242 ± 130 fL (
In this paper, we propose and experimentally demonstrate that the QPI techniques provide unique advantages for the study of individual phytoplankton. High-resolution 3-D RI maps, 2-D synthetic aperture images, and emulated DIC images of live phytoplankton are quantitatively and precisely measured. In particular, we show that from the measured quantitative phase images, the structure and biochemical parameters, including phytoplankton volume, total dry mass, and cytoplasmic density are simultaneously retrieved. Furthermore, internal structures of phytoplankton are clearly visualized without using exogenous labeling agents.
QPI techniques, with their capacity for 3D RI imaging, their potential for generating contrast by intrinsic optical property and their quantitative and non-invasive nature, would be a welcome complement to the existing imaging tools for the study of diverse aspects in phytoplankton biology and biophysics; (1) QPI techniques could be useful for investigating precise morphological and compositional alterations in phytoplankton under diverse environmental conditions, because the RI distributions can serve as an intrinsic optical indicator reflecting the structural and biochemical information. (2) Even though the particular focus of this study is on static 3-D images of phytoplankton, dynamics of 3-D RI tomograms of individual phytoplankton can allow to measure intracellular dynamics of subcellular organelles [39-41] or dynamics fluctuation in cell membrane [42-47] which can provide abundant information about pathophysiology of phytoplankton. (3) In addition, recent advances in QPI techniques can also be further employed to investigate phytoplankton research including super-resolution imaging [48], Fourier transform light scattering technique [49-53], real-time visualization of 3-D RI maps [54], multi-spectral QPI [55-59], and polarization-sensitive QPI [60, 61]. (4) Furthermore, QPI techniques can be accessible to an existing microscope by attaching the recently developed QPI unit [62, 63], which will further extend the applicability of QPI techniques for the study of phytoplankton.
With QPI techniques, many interesting open questions in phytoplankton biology are now accessible to direct experimental study. Furthermore, the present approach can also be extended for the study of general plankton [17, 64, 65] or zoospores of alga [66]. This new approach has many potential applications. For example, when combined with underwater electronics, 3-D RI imaging of live phytoplankton could be of particular interest to marine microbiologists trying to study biological oceanography [67] and plankton ecology [68]. In addition, 3-D RI tomogram of individual phytoplankton can also be utilized for automated plankton identification [69] or the study of fluid dynamics of plankton [62, 70, 71].