The water deer,
According to morphological studies (Allen, 1940; Won, 1967; Won and Smith, 1999), only the Korean subspecies has been recognized in South Korea to date. However, a recent analysis of the mitochondrial
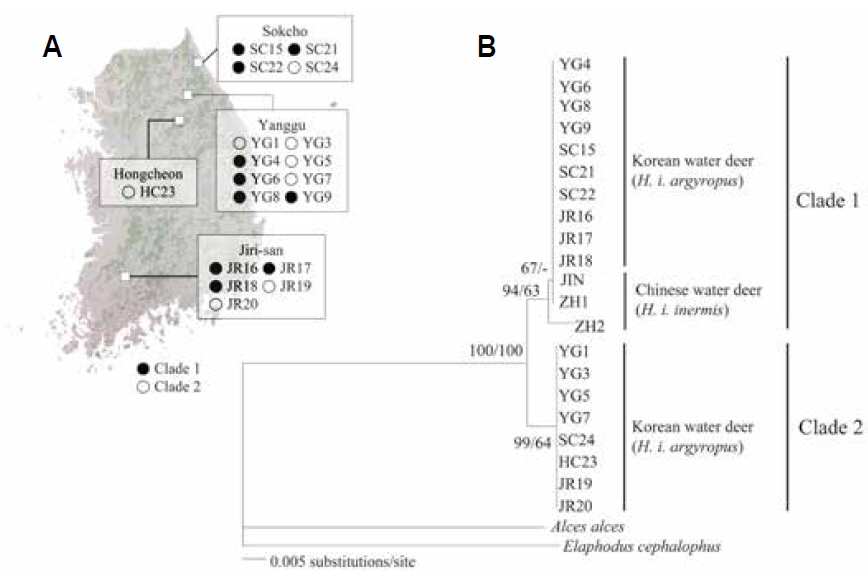
The objective of the present study was to confirm the presence of the two genetic lineages in the Korean populations by analyzing mitochondrial cytochrome oxidase I (COI) gene sequences. We analyzed the intraspecific relationships in Korean water deer by using tissues collected from 18 road-killed individuals from two northern forests (SC: Sokcho, YG: Yanggu and HC: Hongcheon) and one southern forest (JR: Jirisan National Park) of South Korea (Table 1, Fig. 1A). Genomic DNA was extracted from the tissue samples by using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA), and amplified using the InnoTM Taq DNA polymerase kit (Bookyung S∙M, Korea). Primers for PCR amplification of mitochondrial COI gene was performed were Deer F (5′-TTCATTAACCGCTGATTATTTTCAAC-3′) and Deer R (5′-CACGATATGAGAAATTATACCAAACC-3′). This amplification procedure yielded 680-bp fragments of the COI gene; these were aligned using the Clustal-X program package (Thompson et al., 1997) and manually edited using SeA l 1.0a (Rambaut, 1996).
[Table 1.] List of the samples used in this study

List of the samples used in this study
Intraspecific phylogeny was inferred by neighbor-joining (NJ) implemented in PAUP* 4.0b10 (Swofford, 2003) and Bayesian inference (BI) using MrBayes 3.2 (Ronquist et al., 2012). The NJ tree was generated using the K2P model, and the robustness of the tree was tested with 1,000 pseudoreplications. For the BI analysis, the HKY model was selected as the best-fit model under the Akaike Information Criterion(AIC) by using Modeltest 3.7 (Posada and Crandall, 1998). The BI analysis was performed using the Markov chain Monte Carlo technique (MCMC) and one sample every 500 generations was used. The first 25% from each run was discarded as burn-in.
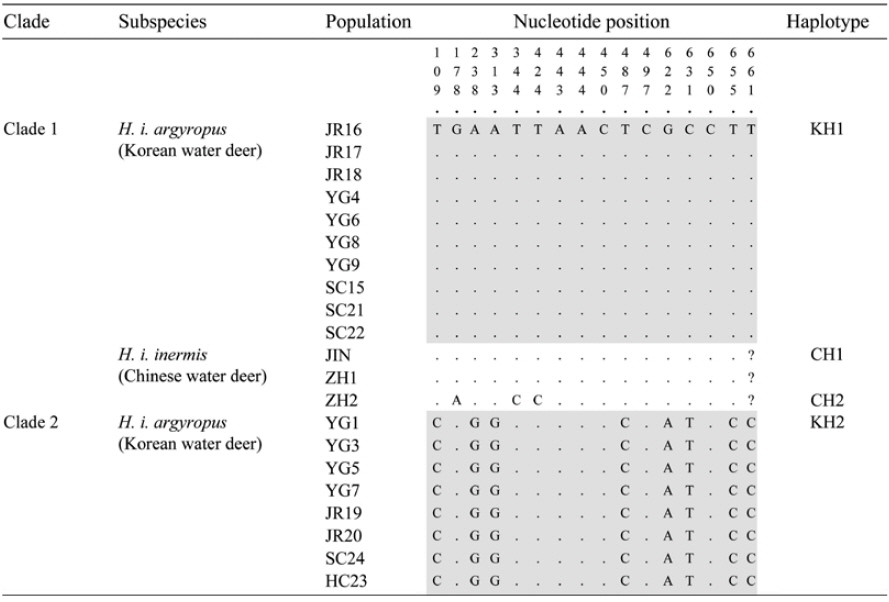
The NJ and BI analyses generated similar trees with identical topologies, with greater support for the NJ tree than the BI tree. The analyses confirmed that Korean water deer could be separated into two phylogroups (Fig. 1B), one of which clustered with the Chinese water deer (JIN, ZH1, and ZH2). This result is consistent with that of a previous study that analyzed the mitochondrial cytochrome
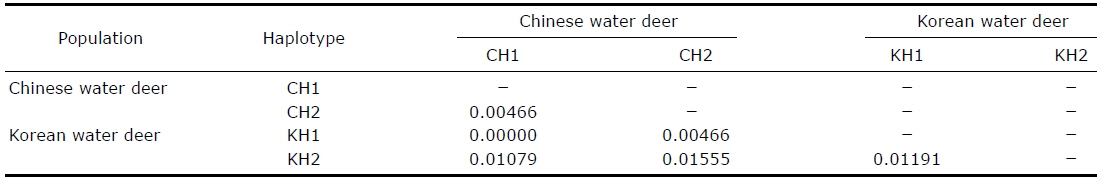
[Table 2.] Pairwise genetic distances between the Korean and Chinese water deer haplotypes
Pairwise genetic distances between the Korean and Chinese water deer haplotypes
We conclude from our intraspecific phylogeny and haplotype analysis based on the mitochondrial COI gene sequence that both the Chinese and Korean water deer are distributed across South Korea. Further morphological studies of the Korean water deer will be required to confirm this reassessment of its taxonomic status.