Diversification rate shifting patterns over evolutionary history have been observed in many taxa (Barraclough and Vogler, 2002; Rabosky et al., 2007; Rabosky and Lovette, 2008a; Alfaro et al., 2009). According to the prediction of adaptive radiation (Schluter, 2000; Losos, 2010; Gillespie, 2013), diversification rates can vary over evolutionary time due to the limitations of ecological opportunities (Ricklefs, 2006; Rabosky, 2009; Harmon et al., 2010; Losos, 2010; Myers and Burbrink, 2012).
Diversification shifting pattern could be modeled in terms of a pattern with multiple-discrete shifts (MDS) (Alfaro et al., 2009; Antonelli and Sanmartín, 2011) or one-continuous shift (OCS) (Rabosky and Lovette, 2008a, 2008b) over the phylogeny. In comparison, a global one-constant-rate model (OCR) predicts that there are no shifting trends of diversification rates over the phylogenetic history, which might be used as a null model when investigating rate-shifting patterns of species diversification (Rabosky, 2007; Rabosky and Lovette, 2008a). MDS and OCR models can be implemented using “MEDUSA” package (Brown et al., 2013), while OCR and OCS models can be performed using “laser” package (Rabosky, 2007) under R environment (R Development Core Team, 2011). There are many systematic comparisons on using both R packages to model rate-changing patterns of species in previous literatures (Alfaro et al., 2009; Fordyce, 2010a; Rabosky et al., 2012). When it comes to the comparison of these two packages, “MEDUSA” package is much more flexible since it could allow multiple rate shifting patterns over the phylogeny (Brown et al., 2013). Thus, alternative hypotheses for testing rate shifting patterns of species diversification can be done easily using this package. In contrast, “laser” package is very limited because only three density- dependent models are implemented (Rabosky, 2006, 2007; Chen, 2013). However, as mentioned above, its advantage is to model the continuous rate shifting pattern.
Amphibian diversity has attracted much attention in recent years due to its high sensitivity to global climate change and habitat destruction (Wake, 1991, 2007). There are many literatures contributing to understand global macroevolutionary and macroecological patterns of amphibians (Grenyer et al., 2006; Buckley and Jetz, 2007; Roelants et al., 2007; Wiens, 2007; Qian, 2009; Pyron and Wiens, 2011; Fritz and Rahbek, 2012). Moreover, diversification analysis of specific or overall amphibian clades has been found in some recent publications (Roelants et al., 2007; Wiens, 2007; Wollenberg et al., 2011; Zimkus et al., 2012). However, diversification rate analysis by comparing alternative rate-shifting models has not been performed on all the living amphibians yet seemingly in comparison to birds (Jetz et al., 2012) or other taxa (Rabosky et al., 2012).
In the present study, by comparing OCS, OCT and MDS models as described above, I evaluate the most appropriate evolutionary model for quantifying global amphibian temporal diversification patterns through model selection and comparison. I implement different models using both “MEDUSA” and “laser” packages.
For the present study, a full dated phylogenetic tree for global amphibians was obtained from the attached data sets in the “geiger” package (Harmon et al., 2008) under R environment (R Development Core Team, 2011), which was reconstructed on the basis of some previous studies (Roelants et al., 2007; Pyron and Wiens, 2011). More information about this tree file is available at URL: http://www.inside-r.org/ packages/cran/geiger/docs/amphibia.
The γ statistic (Pybus and Harvey, 2000) was applied firstly to test whether diversification rates of amphibians would keep constant over evolutionary time scale. If the constantrate assumption was broken down, then time-varying diversification rate models should be employed to fit the phylogeny and the best one was chosen using Akaike Information Criteria (AIC) values.
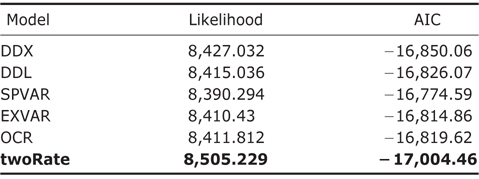
OCS, OCT, and MDS models were fitted onto the amphibian phylogeny using “laser” package (Rabosky, 2007; Fordyce, 2010a). In specific, for OCS models, four models were implemented: DDX, DDL, SPVAR and EXVAR. DX and DDL fit exponential and logistic variants of the density-dependent speciation rate model respectively (Rabosky, 2006, 2007). SPVAR model is a model with an exponentially declining speciation rate through time and constant extinction, while EXVAR is a model with exponentially increasing extinction and constant speciation (Rabosky, 2006, 2007). For MDS models, the two-rate model (coded as “twoRate”) is computed in “laser” package. MDS models with multiple discrete rates were implemented using “MEDUSA” package (Brown et al., 2013). The best number of discrete rate shifts in MDS model was selected through a stepwise AIC procedure(Brown et al., 2013).
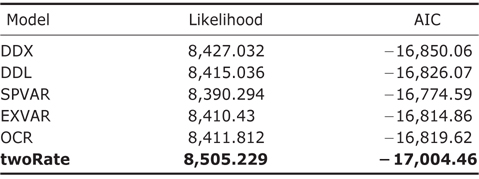
Model comparison and selection of different rate-varying models and the constant-rate model for the diversification rates of global amphibians using “laser” package
When there was no rate shift in the species phylogeny, MDS model was equivalent to OCT model when using “MEDUSA” package. In other words, MDS models nesetd OCT model in “MEDUSA” package. Besides, since OCT is a constant birth-death model, and therefore can be also estimated using the function “birthdeath” in the classical R phylogenetic package “
The γ statistic rejected the null model which stated that there was a globally constant diversification rate of amphibians over the evolutionary time (γ=−5.556, p<0.001). As a consequence, rate-shifting models can be applied to the phylogeny of global amphibians with reasonable grounds.
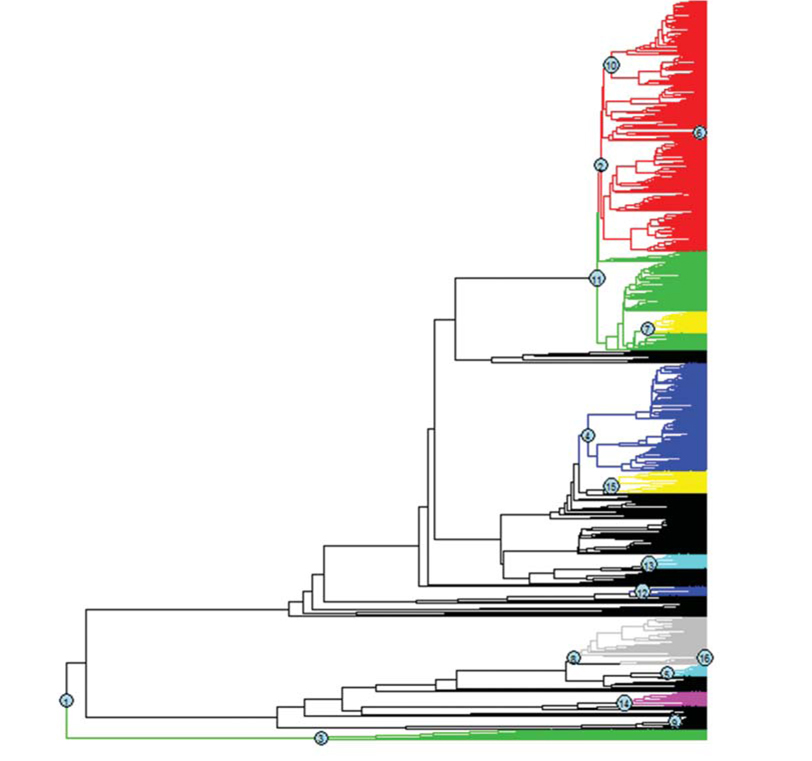
Throughout the comparison of AIC values for different rateshifting models implemented using “laser” package (Table 1), it was found that twoRate discrete model was the best one to characterize diversification rates of amphibians over the phylogeny in comparison to other OCS models. Furthermore, for the MDS models implemented using “MEDUSA” package, it was identified that there were sixteen shifts over different clades in the phylogeny (Fig. 1), further supporting the MDS model.
OCS diversification rate models have been applied and examined by a variety of species assemblages (Rabosky, 2006, 2009; Phillimore and Price, 2008; Rabosky and Lovette, 2008a). MDS models are also growingly applied in many recent studies (Alfaro et al., 2009; Santini et al., 2009; Pyron and Burbrink, 2012; Rabosky et al., 2012; Brown et al., 2013) given its high flexibility on modeling rate shifting patterns and identifying rate-changing lineages using stepwise AIC procedure (Alfaro et al., 2009). For global amphibians, it was found that there were sixteen shifts of the diversification rates in internal nodes over the evolutionary time, suggesting that diversification rates of global amphibians follow MDS models.
“MEDUSA” package is the preferential choice in the analysis of time-varying diversification of species given the fact that multiple shifts of diversification rates are widely observed across different taxa (Jetz et al., 2012; Rabosky et al., 2012) and the rates after shifts can be either accelerated or reduced (Fordyce, 2010b; Jetz et al., 2012). However, “laser” package should be used when one wants to model time-declining OCS situation. But for large phylogenies, multiple shifts should be ubiquitous and thus MDS models may be one of the first choices to implement.
Sampling issue can be a prevailing problem limiting the implication of the present study. This is because phylogenetic trees usually can not cover all the external living species and may result into biases of the results (Chen, 2013). For my study, the amphibian phylogenetic tree used currently contains only 2,871 species, while global amphibians species has a number of over 7,000. Thus, no more than a half of the amphibian species can be represented and subsequently the robustness of the results should be verified in the future when more amphibian taxa are covered in the tree.