Thais Roding, 1798 (commonly known as rock-shell) is one of the most frequently found genera of gastropods worldwide on intertidal rocky shores including those of Japan, China, Taiwan, and Korea. The genus contains about a hundred species names (Houart and Gofas, 2010), some of which includes predatory species that cause serious damage to the oyster culture industry by drilling through the oyster shells (Clench, 1947; Brown and Richardson, 1988). The species of this genus also play an important role in the marine macrobenthic community. They have often been used as an effective indicator species in many marine environmental studies related to human-induced pollution, such as that from organotin compounds (Shim et al., 2000; Hung et al., 2001; Tang and Wang, 2009) and long-term exposure to heavy metal contamination (Rubio et al., 1993; Han et al., 1997; Blackmore and Wang, 2004).
Despite their significance in aforementioned environmental studies, the species-level taxonomy of this group is quite complicated due to morphological variations in shell characteristics (Tan and Sigurdsson, 1996). Indeed, the morphological features used most frequently for species identification, including size, shape and the absence or presence of blotches on the nodules of shell surfaces and shell apertures, are highly variable depending on the local environment (Hayashi, 1999; Tan and Liu, 2001). Owing to the substantial variability in shell characteristics, species identification relying on shell morphology alone often leads to erroneous results creating many synonyms. As a result, the associated nomenclature for Thais species is unusually complicated (Clench, 1947; Tan, 1995).
In Korea, Thais species have been included as part of a species checklist in the faunal report on mollusks. Earlier studies, including some encyclopedias of Korean shells, relied entirely on the shell characteristics without careful appraisal of species identity (Choe, 1992; Lee and Min, 2002; Min, 2004). A taxonomic investigation of Korean muricid species based on a morphological comparison of shell and radula characteristics showed that three Thais species are very abundant in Korean coastal areas (Choe and Park, 1997): T. clavigera (Kuster, 1858), T. bronni (Dunker, 1860), and T. luteostoma (Holten, 1803). In contrast to taxonomic assessments by most earlier authorities, these species are not very morphologically discernible from each other because their shell morphology is greatly variable, creating a diverse array of local morphotypes according to their geographic origins. Therefore, there is considerable taxonomic confusion with regard to species-level taxonomy in this group.
The utility of molecular markers in modern taxonomy has been growing rapidly as a reliable tool for examining the phylogenetic relationships in different levels of taxonomic groups and species identification. As in many other animal groups, molecular data from mitochondrial (mt) gene fragments (e.g., 16S and cox1) provide a wealthy resource for assessing the genetic variations and phylogenetic signals among closely related species of diverse molluscan taxa (Boudry et al., 2003; Park and Kim, 2003; Lam and Morton, 2006; Reece et al., 2008). The present study examined the genetic variations and relationships among three Korean Thais species based on partial nucleotide sequences of mt cox1 gene fragments. In addition, the sequence data presented here provides a very useful molecular identification tool for the three Korean Thais species, which are often taxonomically complicated when based upon the morphological characteristics alone.
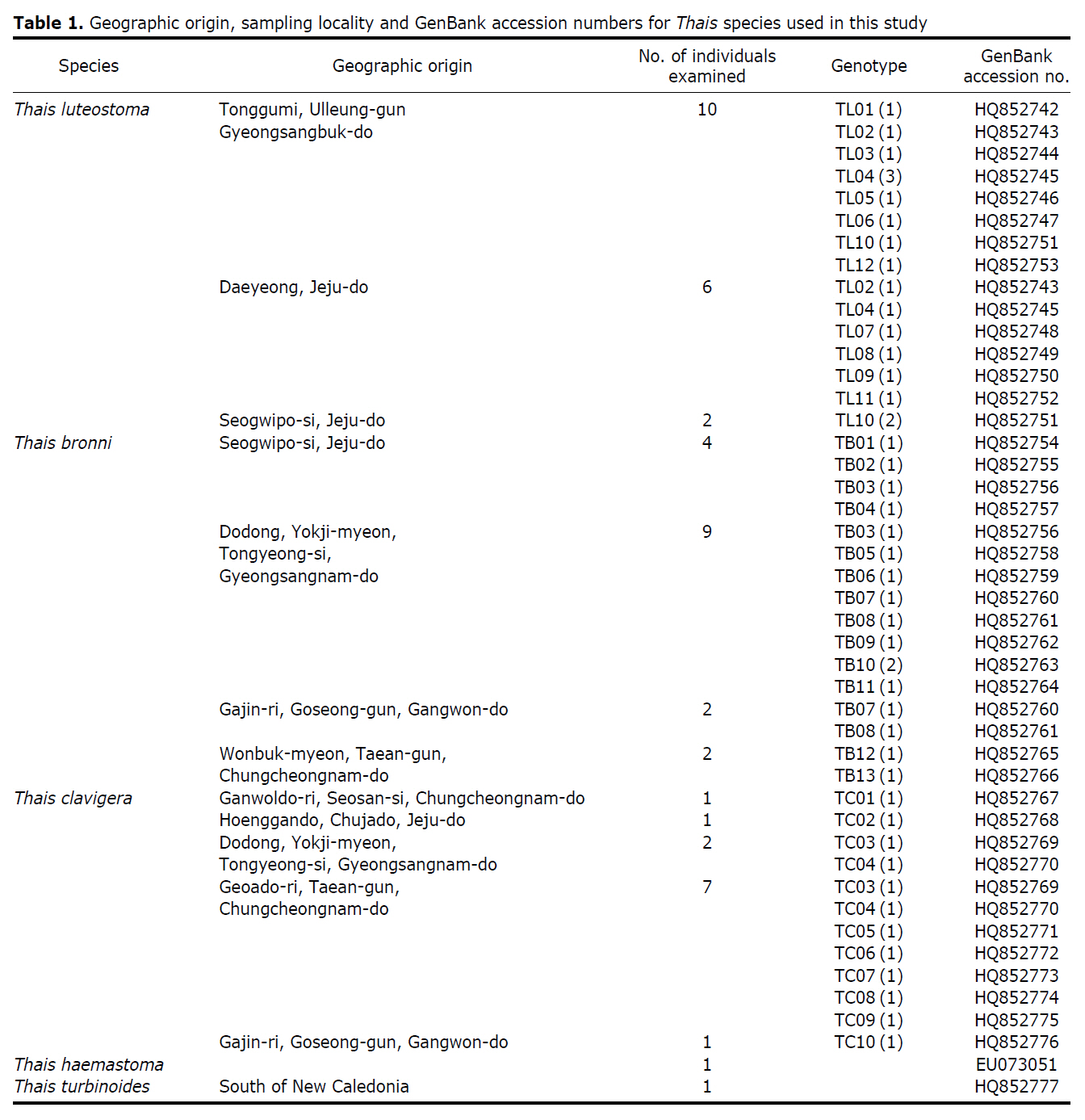
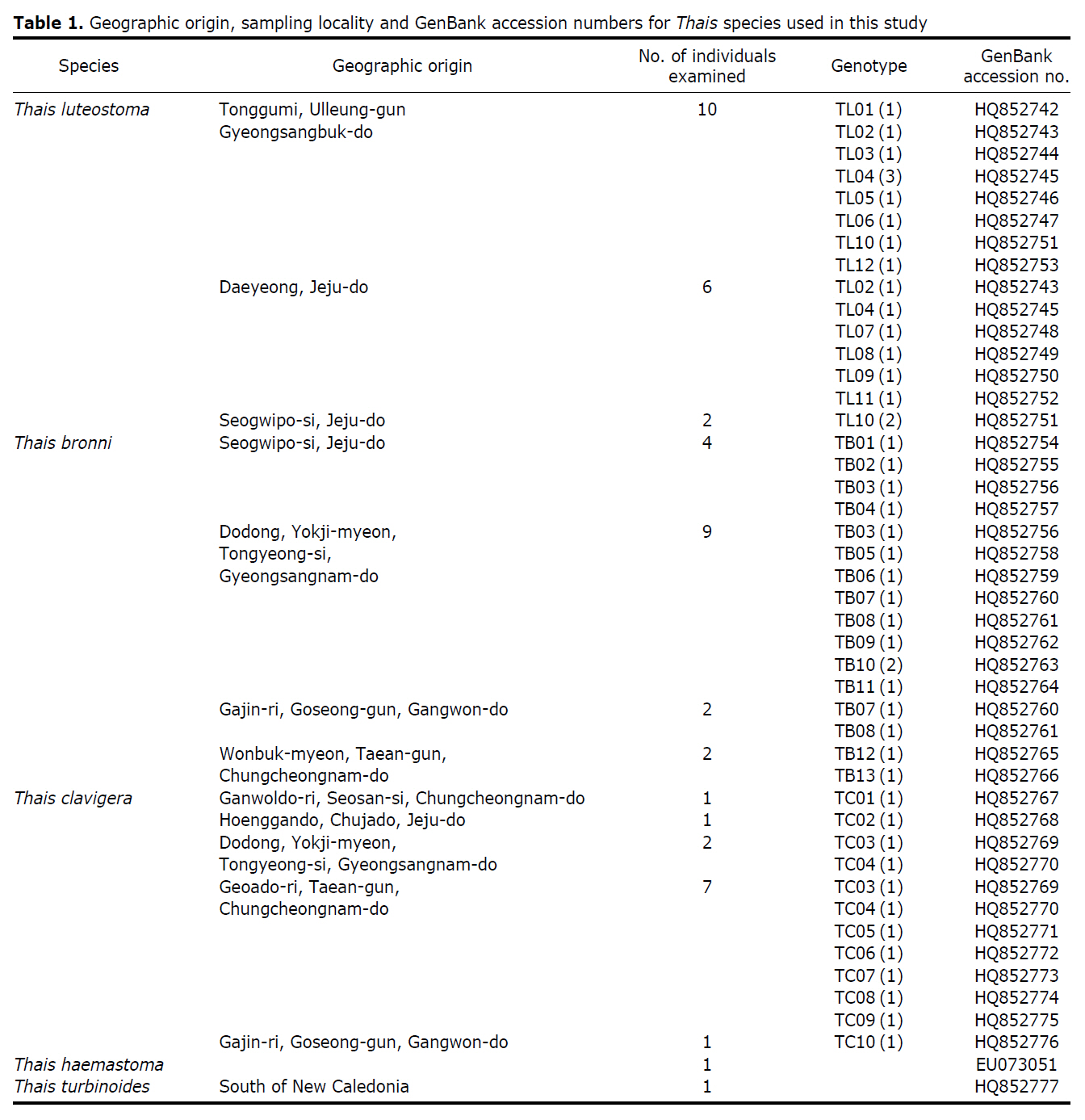
The specimens were collected from the intertidal and/or subtidal zones in nine localities of the Korean seashore. Voucher specimens used for species identification and subsequent molecular sequencing were deposited in the Marine Mollusk Resource Bank of Korea (MMRBK; Chungbuk National University, Korea). Identification of the Thais specimens was carried out based on the shell characteristics, from which they were assigned to three species: T. clavigera, T. luteostoma, and T. bronni (Fig. 1). A total of 46 specimens sampled from nine localities representing three Korean Thais species were used for sequencing analysis. The total genomic DNA was extracted using a tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. A universal primer set (LCO1490: 5'-GGTCAACAAATCAT AAAGATATTGG-3', HCO2198: 5'-TAAACTTCAGGGT GA CCAAAAAATCA-3') was employed for PCR amplification of the target gene fragment mt cox1 (Folmer et al., 1994). PCR reactions were performed in a 50 μL reaction volume consisting of 10 units of Taq polymerase (Roche, Mannheim, Germany), 2.5 mM dNTP mixture, 2.5 mM MgCl2 and 20 pmole of each primer with the following amplification conditions: one cycle of the initial denaturation step at 94℃ for 2 min, followed by 35 cycles of denaturation at 94℃ for 30 sec, primer annealing at 45℃ for 30 sec and elongation at 72℃ for 1 min with a final extension step at 72℃ for 10 min. The PCR-amplified target gene fragment was purified using a QIAquick gel extraction kit (Qiagen) according to the manufacturer’s protocol. The sequencing reaction was performed using a BigDye® terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA), and the reaction products were electrophoresed using an ABI 3730XL DNA analyzer. Phylogenetic analysis for the mt cox1 dataset obtained from the three Thais species was performed using different tree-building methods (maximum parsimony [MP], neighbor-joining [NJ], and maximum likelihood [ML]) with PAUP 4.0b10 (Swofford, 2002) using Thais turbinoides as an outgroup. NJ and ML analyses were carried out using the HKY+Γ (0.0913 of gamma shape parameter) model that was selected as the ‘best-fit’ substitution model according to the Akaike information criterion (AIC) from implementation of the Modeltest version 3.7 program (Posada and Crandall, 1998). The reliability of grouping in both MP and NJ methods was estimated using non-parametric bootstrap resampling of 1,000 pseudo-replicates.
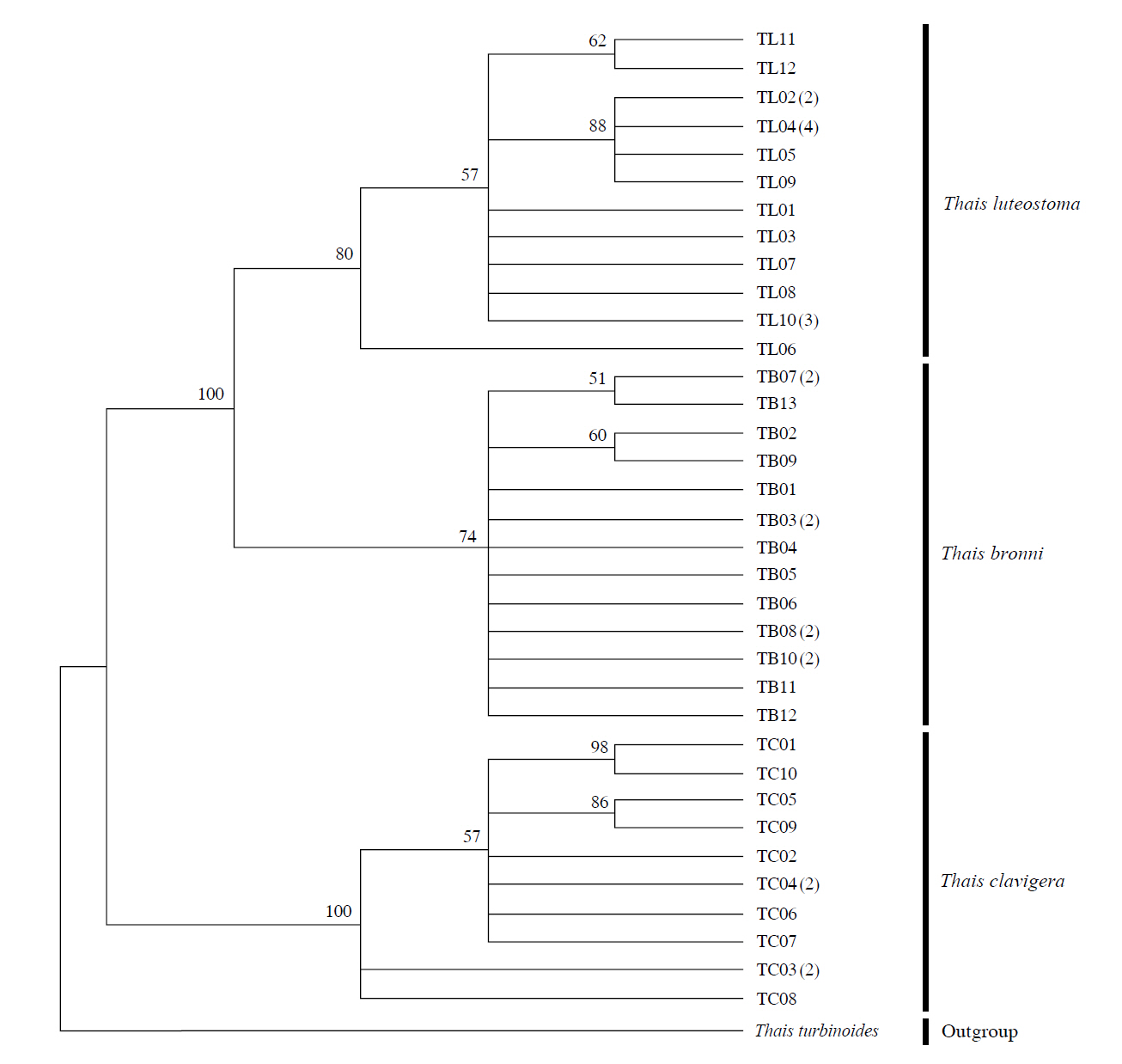
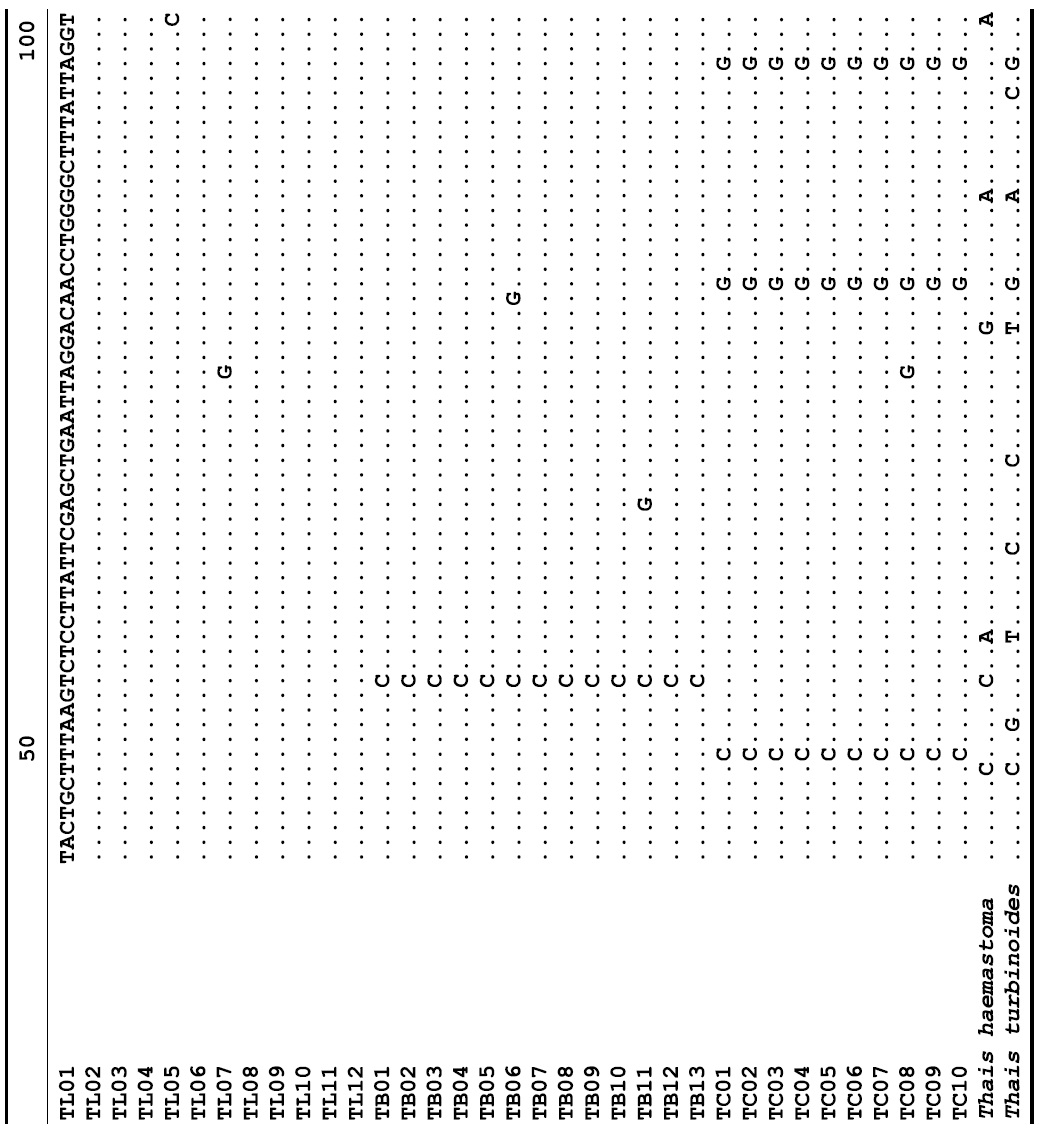
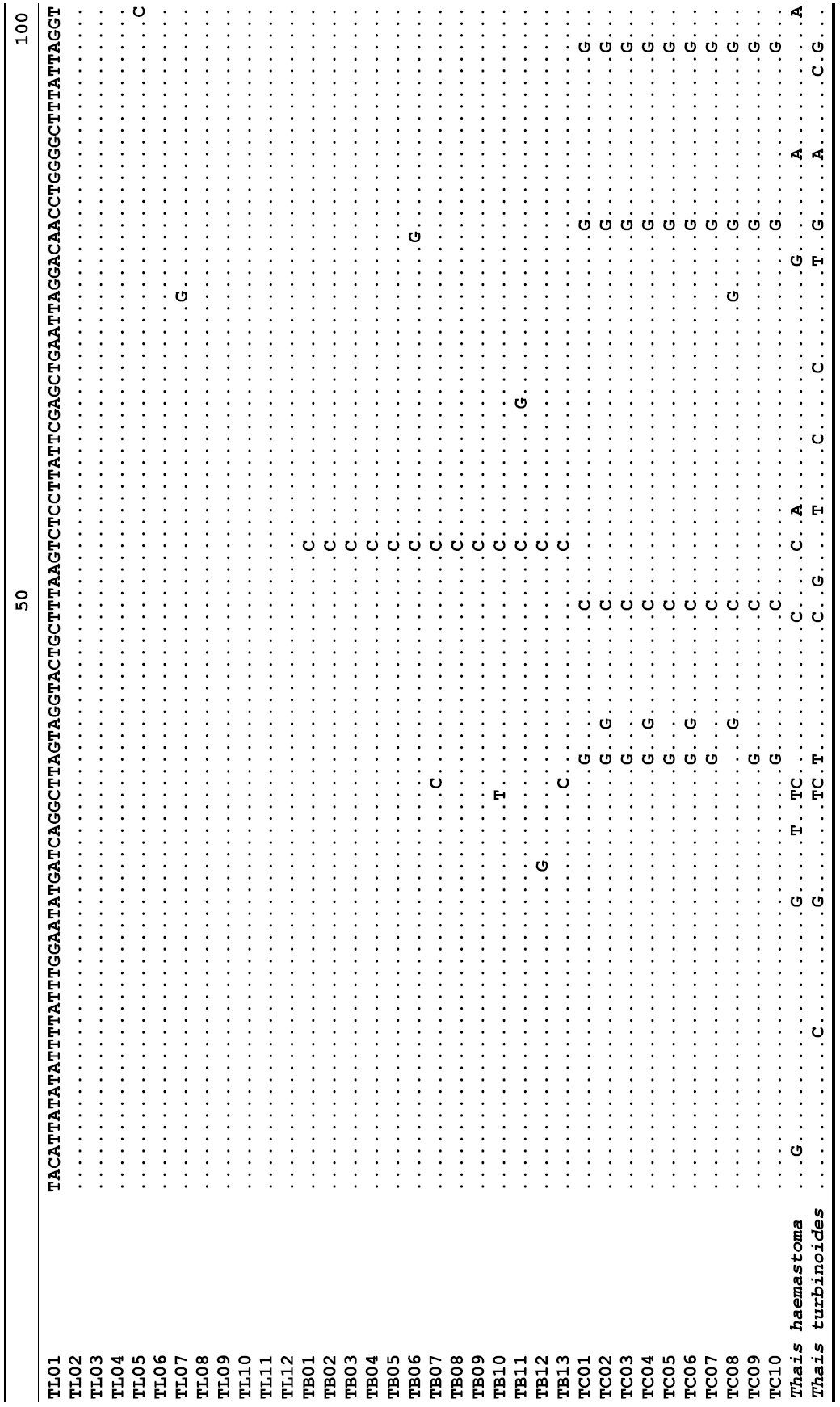
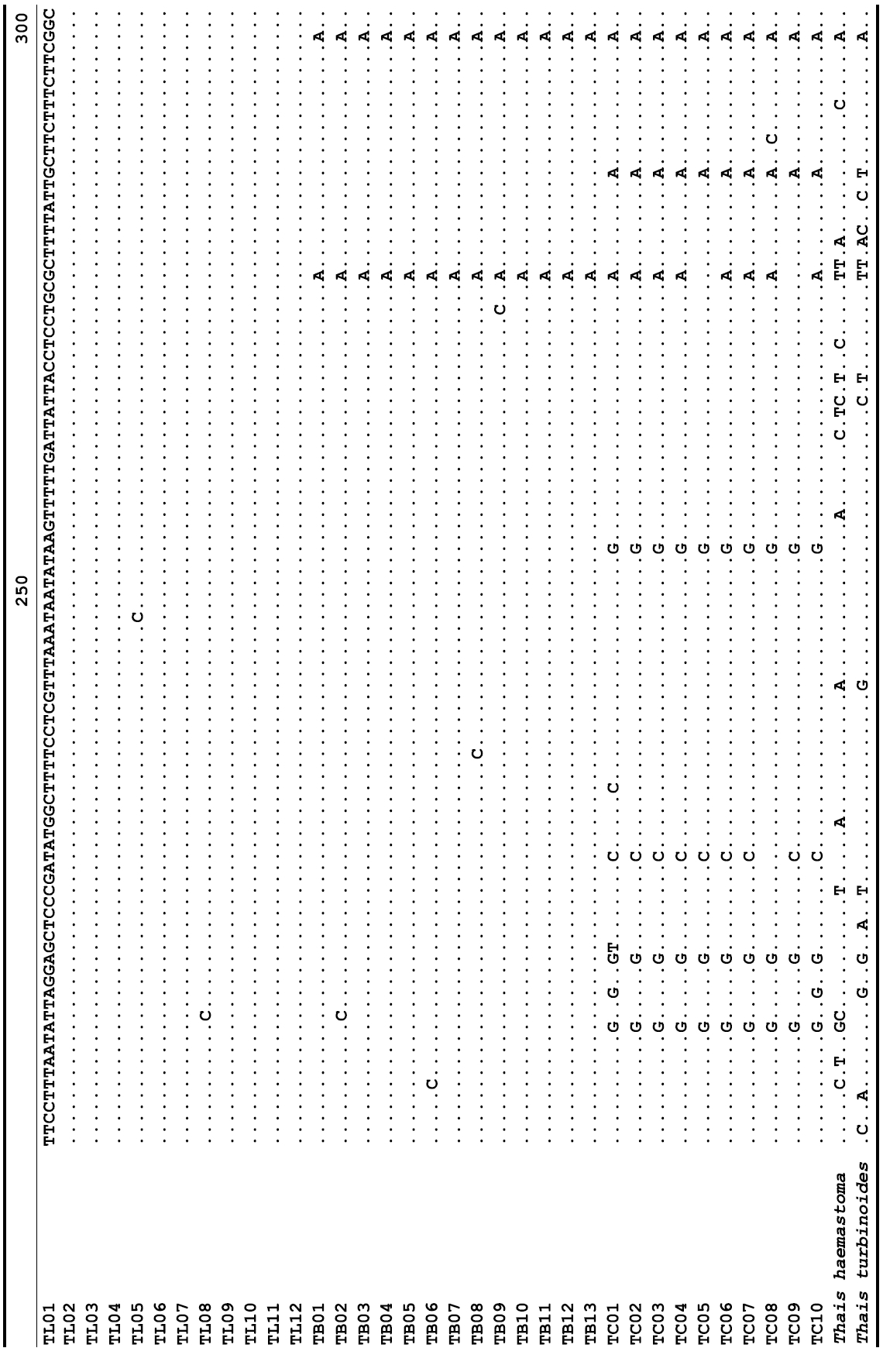
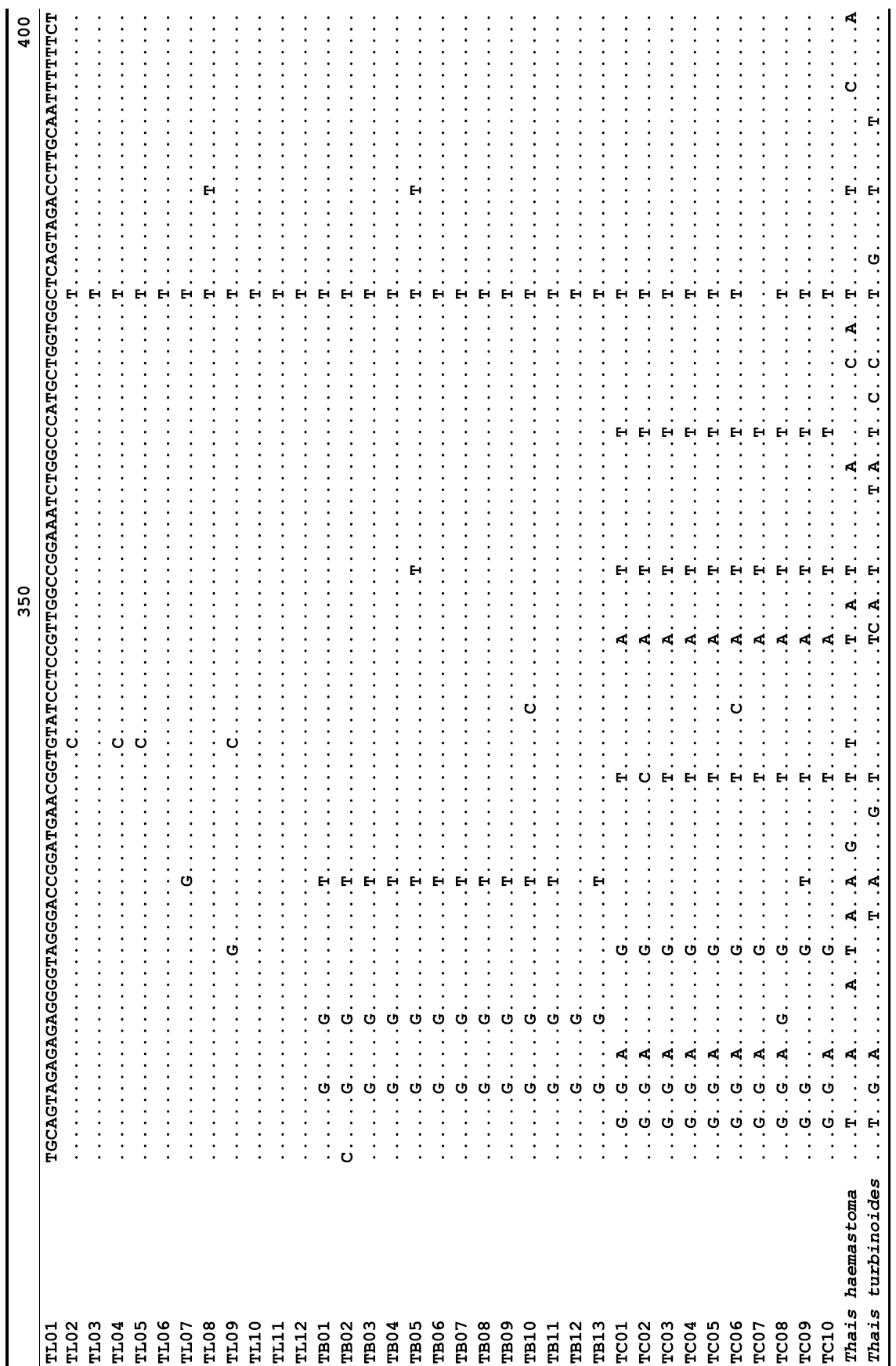
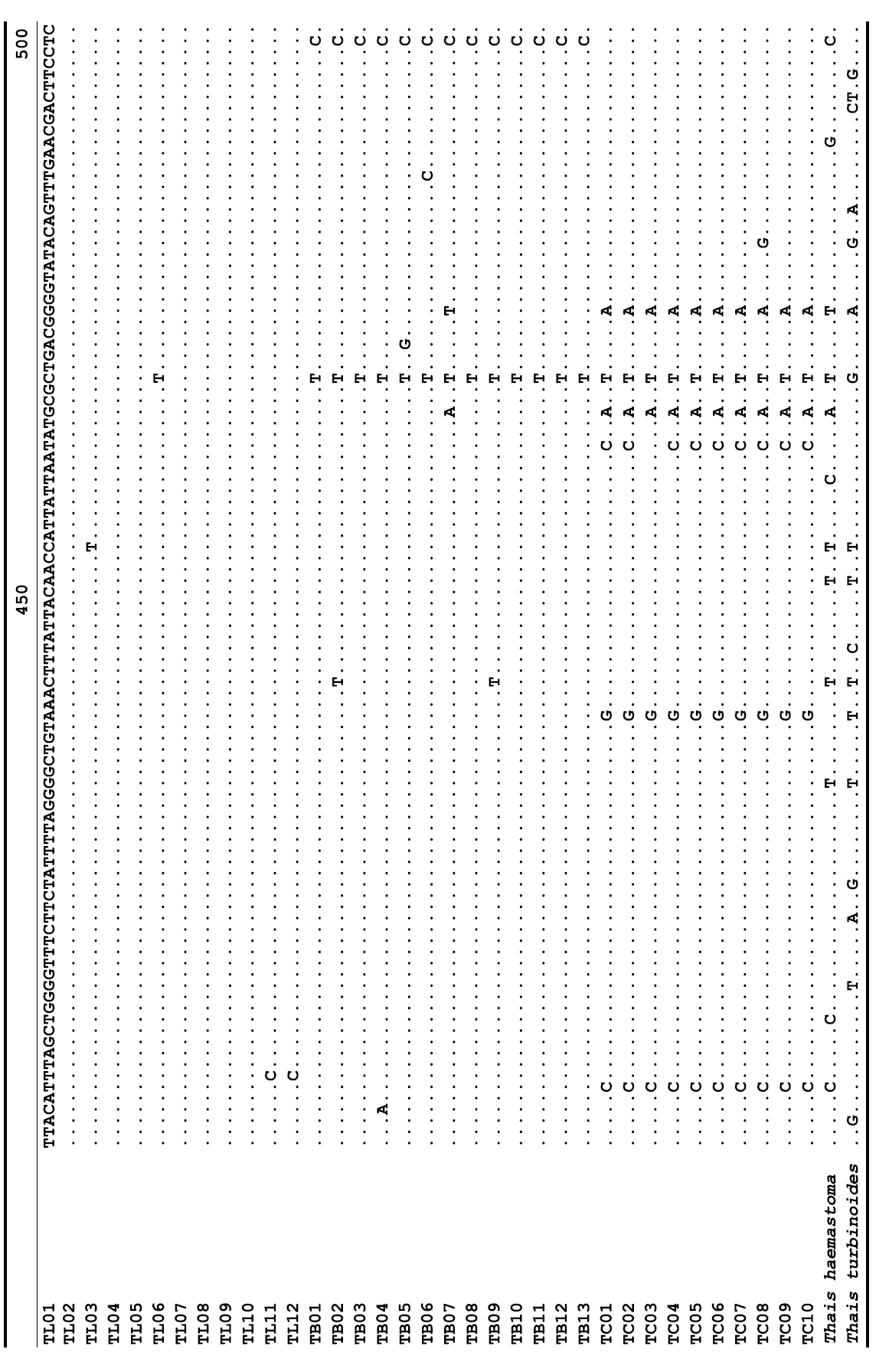
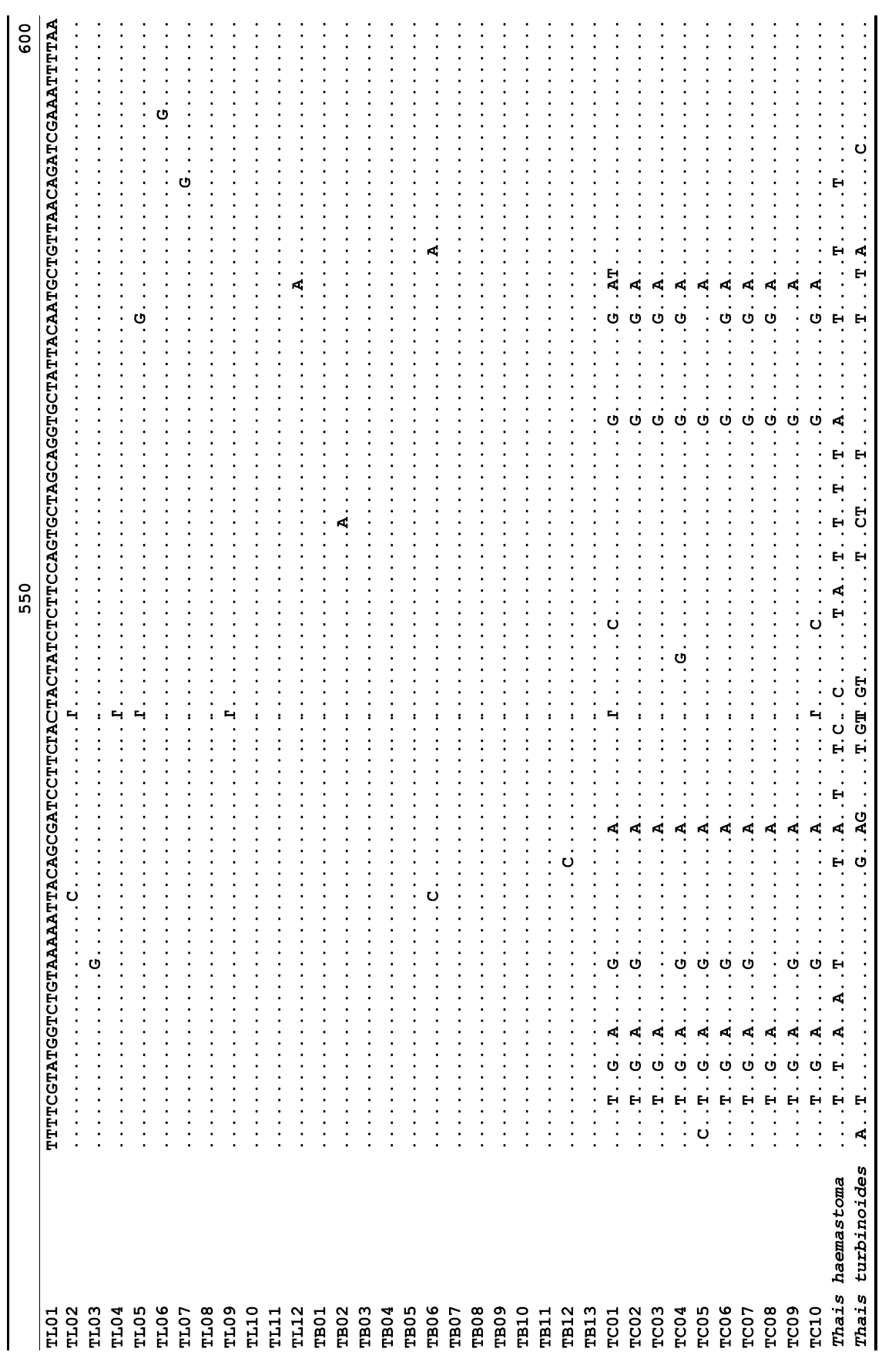
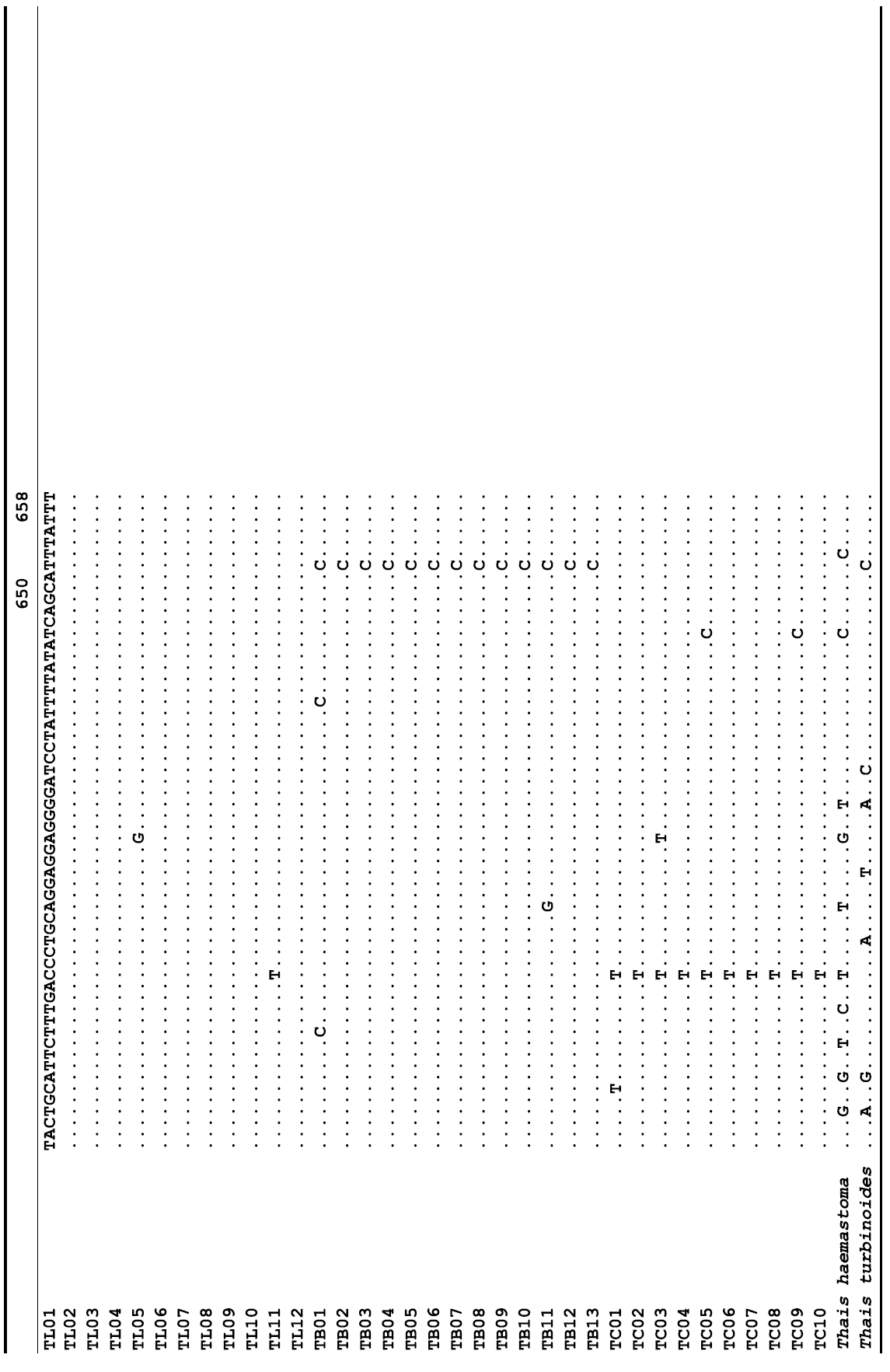
The target mt cox1 gene fragment determined for all Thais species is 658 base pairs (bp) in length with no intra- or interspecific length variations. Table 1 lists the GenBank accession numbers for each mt genotype and locality data for the species. The MP analysis of the mt cox1 dataset showed two distinct clades with very strong support (Fig. 2): one representing the assemblage of mt genotypes from T. clavigera and the other containing two groups of mt genotypes,
representing T. luteostoma and T. bronni. The grouping patterns among the three species in the MP analysis were identical to those of the NJ and ML analyses (not shown). The bootstrap values for these two clades were very high (100% for T. clavigera in both MP and NJ methods, and 96% and 100% for T. bronni-T. luteostoma in MP and NJ analyses, respectively). The branches representing T. luteostoma (92% and 96% bootstrap supports in MP and NJ, respectively) and T. bronni (88% and 81% supports in MP and NJ analyses, respectively) also received relatively strong support. In contrast to the well-structured relationships, the inferred tree showed no particular grouping pattern among the mitochondrial genotypes within each of the three species. Irrespective of the analytical methods (MP, NJ, and ML), the reconstructed phylogenetic trees consistently depicted T. bronni and T. luteostoma as being more closely related to each other than to T. clavigera. This sister group relationship was robustly supported by a bootstrap value of 96% (MP) and 100% (NJ)
in the different analytic methods (MP and NJ). This result corresponds to the morphological similarities revealed by some earlier studies using comparative analysis of morphological characters (shell, radula, and penis morphology etc.) (Choe and Park, 1997; Tan and Liu, 2001).
Aside from the shell external characteristics, traditional taxonomy has appreciated the radula morphology as one of the most useful features for the classification of a wide range of gastropod groups, including muricid species (Cooke, 1919; Wu, 1965). Nevertheless, the radula characteristics are often quite similar among closely related congeneric species, making species identification rather difficult based on the radular traits alone (Fujioka, 1985; Tan and Liu, 2001). Some earlier authorities reported that the rachidian teeth of T. luteostoma and T. bronni were quite similar but noticeably different from those of T. clavigera in both the shape and number of denticles. There are usually more than six denticles between the lateral and marginal cusps recorded for the Korean population of T. luteostoma and T. bronni (6 and 7-8, respectively) (Choe and Park, 1997). Tan and Liu (2001) observed some variations from Hong Kong population of T. luteostoma in the denticle number, ranging from five to eight. The rachidian tooth of T. clavigera has fewer denticles (normally 3-4 denticles between lateral and marginal cusps). Although variations in radula characteristics according to the local populations are not uncommon, particularly in the denticle number, similarities in the radula characteristics between T. bronni and T. luteostoma are in line with the close relationship uncovered by genetic analysis of the molecular data in this study. The closer relationship between T. bronni and T. luteostoma is also supported by a comparison of the penis morphology. Indeed, the morphology of the male reproductive organ is generally considered speciesspecific and appears to be very useful for species identification in this genus. Exceptions were found for T. luteostoma and T. jubilaea, whose penis morphology was more or less similar (Tan and Sigurdsson, 1990; Tan and Liu, 2001). Interestingly, the penis morphology of T. luteostoma was quite different from that of T. clavigera, but similar to that of T. bronni from the Korean population. T. bronni has a recurved hooklet-shaped penis with a gradually tapered terminal end (Park, 1996) and these characteristics were also found in Hong Kong (Tan and Liu, 2001) and Korean (personal observations) populations of T. luteostoma. The morphological similarities of both radula and penis are consistent with the current molecular data, indicating that T. luteostoma and T. bronni are very closely related. The inclusion of broader taxon sampling will be needed to place the Korean population of species within the phylogenetic framework of the genus.
A total of 35 mt genotypes were discovered from 46 individuals of nine local populations of three Korean Thais species (Aligned cox1 sequences for Thais species are shown in Supplementary data, Appendix 1). Each of these three species included a large number of mt genotypes: 10 for T. clavigera, 13 for T. bronni and 12 for T. luteostoma. Table 2 lists the uncorrected pairwise (p) distance among mt genotypes. The sequence divergence of T. turbinoides (outgroup species) from Korean species ranged from 15.2% (between TL08 of T. luteostoma and T. turbinoides pair and between TB07 of T. bronni and T. turbinoides pair) to 16.9% (between TC01 of T. clavigera and T. turbinoides pair). The highest interspecific sequence divergence (17.7%) was observed between T.haemastoma and the genotype TL11 from T. luteostoma. On the other hand, sequence comparison within Korean congeneric species revealed the T. clavigera sequences to differ considerably from those of T. luteostoma and T. bronni, with maximum differences ranging from 7.14% (between TC01 and TL07 pair) to 7.75% (between TC01 and TB06 pair). In contrast, interspecific sequence divergence between T. luteostoma and T. bronni was relatively very low, ranging from 1.52% (for TL10 and TB03 pair) to 3.19% (for TL05 and TB06 pair). The lowest interspecific difference value (1.52%) detected between T. luteostoma and T. bronni was even lower than the highest divergence value detected within T. clavigera (2.43% between TC01 and TC08). This low divergence value found between T. luteostoma and T. bronni implies that cladonegenetic split of these two species is very young with a very recent common origin. Further molecular survey using different molecular marker is required to fully understand a detailed picture of the origin for their low level of sequence divergence. In addition, sequence comparison of conspecific individuals revealed extensive sequence variations within each of the three species. The intraspecific differences among the T. clavigera mt genotypes were remarkably high, with a maximum 2.43% divergence of a pairwise comparison(found between TC01 and TC08). The sequence divergence within species in each of T. bronni and T. luteostoma was also high: the maximum value was detected between TB06-TB02 and TB06-TB07 pairs in T. bronni (1.37%) and between TL05-TL06, TL05-TL07, and TL05-TL12 pairs in T. luteostoma (1.37%).
Along with the considerable genetic divergence within species, there were a large number of mt genotypes discovered in each of the three species. Thirty five mt genotypes were detected from 46 individuals of the three species sequenced. Of these, a small fraction of mt genotypes (9 of 35; 25.7% of total genotypes detected) is represented by more than one individual (Table 1, Fig. 2). With the exception of these, all other mt genotypes (26 of 35; 74.3% of total genotypes detected) were presented by a single individual. These exclusive mt genotypes were shown to be unresolved polytomies at the terminal tips in the reconstructed phylogenetic tree (Fig. 2). This unexpectedly high genotypic diversity encountered within each of the three Korean Thais species is assumed to be a reflection of the genetic diversity introduced from potential source populations with diverse geographic origins, such as Taiwan, Hong Kong, and many different coastal regions of South China and Japan. Although a large expansion of deep oceanic water is considered an obstacle to the longdistance dispersal of many marine benthic taxa, some coastal invertebrate taxa have crossed this barrier using a prolonged pelagic larval form (Scheltema, 1971, 1986, 1988) or by rafting (passive transportation of sessile-form juveniles attached to drifting objects) (OFoighil et al., 1999). In cases of long range dispersion, the juvenile forms of these animal groups may have the potential to transport and establish successful colonies thousands of kilometers away in downstream habitats that are geographically distant from up-
stream source populations using the oceanic current systems. Indeed, Thais species have a wide distribution range in the Asian Pacific Ocean from the South China Sea area (including Hong Kong, Taiwan) to Northeast Asia (including Japan and Korea) (Kuroda et al., 1971; Choe and Park, 1997; Tan, 2000; Tan and Liu, 2001). It is possible to envisage that a continuous influx of larval immigrants from their potential source populations might contribute to some extent to the near-shore malacofauna of the Korean Peninsula. Of the western boundary currents in the North Pacific Ocean, the Kuroshio Current System is a major component of the most influential warm current that leaves the east coast of Taiwan. Its surface water effects travel northeastward to many regional rocky shores in eastern China, mainland Japan,and the Korean Peninsula by streams of the Tsushima Current and Yellow Sea Current. These current systems running northeastward may be able to transport planktotrophic larvae or drifting objects that carry sessile forms of life (e.g., egg capsules of Thais species) to many different downstream islands and/or continental rocky shores. The routine influx of these types of immigrants from geographically distant source populations with different geographic origins (e.g., Taiwan, Hong Kong, and many different coastal regions of South China and Japan) and their ability to colonize in downstream habitats might act as an effective supplier of new recruitment, thereby shaping the contemporary population structure of Korean Thais species. Under this assumption, the mt genotypic diversity found among Korean Thais populations is likely to reflect a subset of the genetic diversity of their potential source populations. Sampling from their potential source populations was not included in the present study. Therefore, further comprehensive taxon sampling from unstudied potential source populations will be needed to determine the origin and key factors responsible for the high level of genetic diversity discovered within each of the three Korean Thais species examined.