



Identification of endophytic yeasts inhabiting the internal roots of the Mankyua chejuense tree requires techniques involving biotechnology. There is a need for a culture-based method to isolate and identify yeast strains associated with M. chejuense.
We spread homogenized M. chejuense root samples onto glucose-peptone- yeast agar containing antibiotics, Triton X-100, and L-sorbose. A total of 152 yeast isolates were obtained and identified via phylogenetic analysis based on ITS gene sequencing. The results revealed that the root-associated yeast species included the genera Cyberlindnera (140 isolates), Candida (11 isolates), and Kluyveromyces (one isolate). Additionally, three yeast isolates showed high bioethanol production.
We identified the specific yeast community associated with M. chejuense roots. These yeast isolates may have industrial applications as bioethanol producers. Our findings revealed that Cyberlindnera isolates included C. suaverolens and C. satumus, while Kluyveromyces isolates showed high bioethanol production.
Yeast participate in the complex processes within ecosystems such as plant tissues, insects endosymbionts, soil, as well as aquatic and extreme environments (Fonseca and Inacio, 2006; Raspor and Zupan, 2006; Botha, 2011). Industrial applications of yeast include food fermentation (Deak, 2009; Tamang and Fleet, 2009).
In this study, we explored yeast species associated with
[Table 1.] Characteristics of M. chejuense habitat
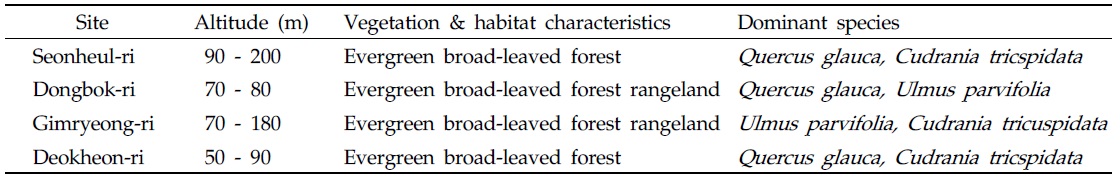
Characteristics of M. chejuense habitat
There is significant research interest in ethanol production by yeast. Recently, non-conventional yeasts (NCY), excluding
>
Yeast isolation from roots of M. chejuense
Yeasts were cultured on agar media in square plates (245 × 245 × 25 mm, Nunc Bio-Assay Dish; Thermo Scientific, Roskilde, Denmark). All colonies from one or two individual plates were picked up and cultured separately. A total of 152 individual isolates were transferred to fresh plates three times and then processed for sequencing of ITS (internal transcribed spacer) genes.
>
Sequencing and phylogenetic analysis of yeast isolates
Sequencing and phylogenetic analysis were performed as previously reported (Choi
>
Phylogenetic analysis of Cyberlindnera isolates
Nucleotide sequences of ITS genes were aligned using the Clustal Omega program on the EMBL-EBI website. The Basic Local Alignment Search Tool (BLAST) search was used to identify GenBank sequences representing the yeast strain most closely related to each isolate. Phylogenetic trees were constructed by the neighbor-joining method, using MEGA5 for Windows (Tamura
Bioethanol production was analyzed from the filtrate obtained after anaerobic culture of 30 random isolates, for 48 hours. Analysis was performed by NICEM (National Instrumentation Center for Environmental Management, Seoul, Korea) by high- pressure liquid chromatography (H-column, Dionex Ultimate3000, USA), using refractive index detector (ERC, Refracto MAX520, Japan) and ultraviolet light at 210 nm.
In this study, wild-type yeast community associated with
We previously showed that
The
[Table 2.] Bioethanol productivity of representative root yeasts associated with M. chejuense
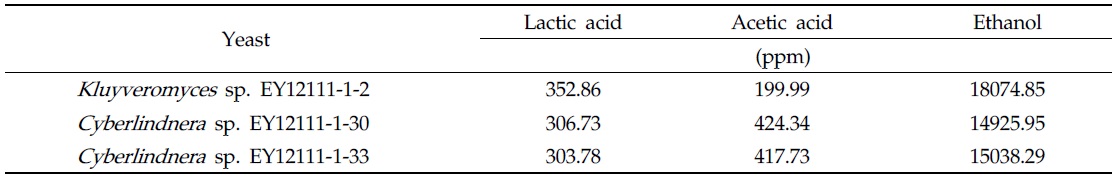
Bioethanol productivity of representative root yeasts associated with M. chejuense
In conclusion, we isolated yeast from roots of an endemic tree,




