In recent screenings of natural products by chemists, more cyclopeptide alkaloids have been discovered in marine organisms than in plants. However, approximately 500 cyclopeptides have been isolated from higher plants during the past half century, and most of these are contained in species from the Caryophyllaceae and Rhamnaceae families.1 Among these, compounds found in Rhamnaceae are known as cyclopeptide alkaloids. The molecules consist of 13-, 14-, or 15-membered macrocyclic ring structures, which are composed of a styrylamine moiety and two or three á-amino acid residues.1-3 According to the sizes of these macrocyclic rings, cyclopeptide alkaloids are categorized into three groups: type Ia, Ib, and Ic.
Seeds of wild jujube trees, Ziziphus jujuba var. spinosa, have been used as treatments for insomnia and anxiety in traditional medicines of Asian countries, including China and Korea.4 Modern studies on the phytochemicals of Z. jujuba var. spinosa seeds suggest that flavonoids,5 saponins,6 and alkaloids7,8 contribute to the sedative effects of the seed extracts. Insomnia and anxiety are common mental disorders, and the personal and social burdens of these psychiatric diseases are considerable.9,10 Therefore, cyclopeptide alkaloids have been studied widely as phytochemicals because of their potential as chemotherapeutic agents for the treatment of insomnia and anxiety disorders.
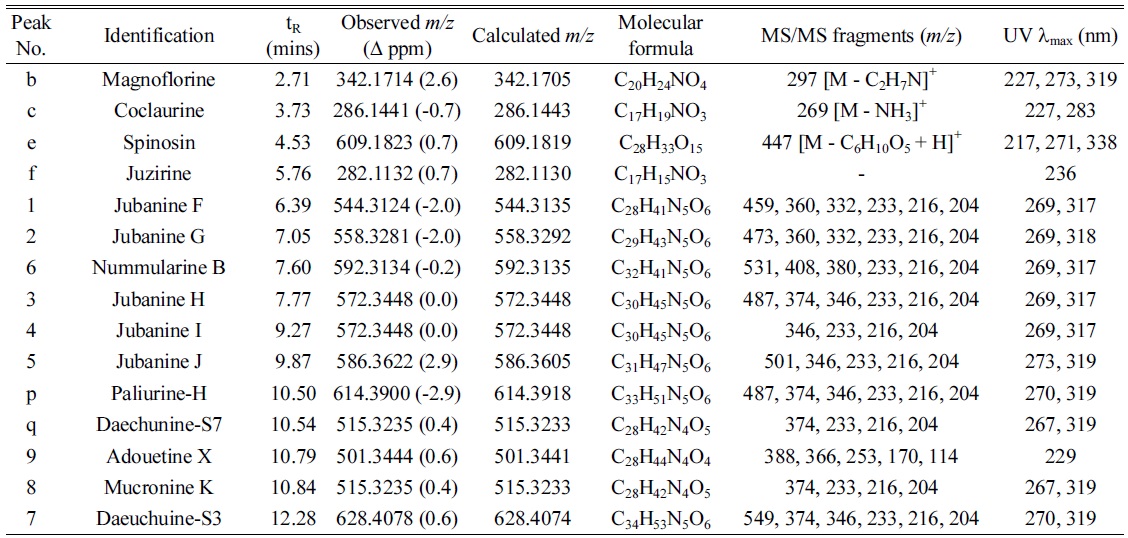
In our last study, five new cyclopeptide alkaloids, jubanines F-J (1-5), were isolated and identified from the roots of Z. jujuba, the most common species of Rhamnaceae, along with three known compounds, nummularine B (6), daechuine-S3 (7), and mucronine K (8).11 On the basis of this study, we realized that there are no established methods for the chemical analysis of these molecules. Modern analytical advances such as UHPLC-ESI-qTOF-MS have enabled many researchers to systematically analyze the complicated matrix of phytochemical compounds. UHPLC provides high-resolution separation of large numbers of constituents,12 whereas the qTOF-MS detector provides structural information from the separated chromatographic peaks.13 Thus, the development of such analytical methods would enhance the scope of future studies on cyclopeptide alkaloids.
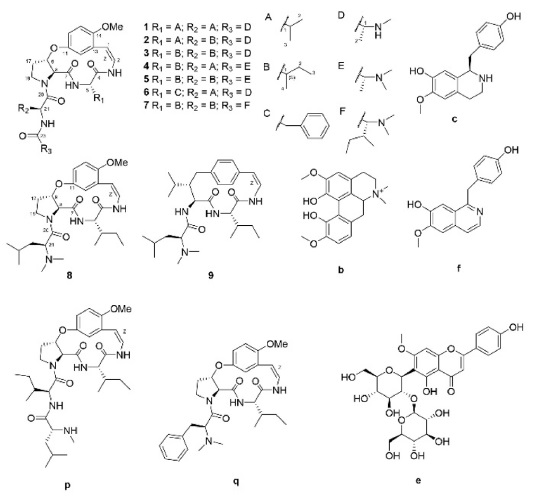
In the present study, an analytical method optimized for Rhamnaceae cyclopeptide alkaloids was developed. In addition to previously isolated type-Ib compounds 1-8, adouetine X (9), a type-Ia cyclopeptide alkaloid compound, was separated from the alkaloid-rich fraction of Z. jujuba root extract. Using these structurally identified compounds and alkaloid-rich fractions of Z. jujuba roots and Z. jujuba var. spinosa seeds, the chromatographic method was optimized, and the MS and MS/MS fragmentation spectra of these compounds were analyzed. MS/MS spectra were obtained using the MSE technique, which is a promising approach for generating molecular fragment information on large numbers of unknown chemicals.14 By investigating the fragmentation pattern of cyclopeptide alkaloids, various unidentified molecules could be tentatively characterized.
HPLC grade water and acetonitrile were purchased from Avantor Performance Materials. Inc. (Central Valley, PA, USA). The alkaloid-rich fraction of Z. jujuba roots and compounds 1-8 were prepared
as described in our previous study.11 Compound 9 (25.1 mg) was additionally isolated from subfraction A2c by preparative HPLC, and its chemical structure was identified by comparing the NMR data with the reference.15 Reference standards of magnoflorine and spinosin were purchased from ChromaDex Inc. (Irvine, CA, USA).
The seeds of Z. jujuba var. spinosa were purchased from the Gyeongdong medicinal herb market in Seoul, Korea. The alkaloid-rich fraction was obtained through an acid-base extraction method. Twenty-one grams of dried and ground Z. jujuba var. spinosa seeds were extracted with MeOH (500 mL) at room temperature under ultrasonication for 4 h to yield 1.3 g of dried extract. The extract was suspended in H2O and acidified with 1N HCl to pH 3.0. The acidic solution was first extracted with EtOAc to remove other hydrophobic constituents. The aqueous residue was basified with 1N NaOH to pH 9.0 and extracted with CHCl3 to yield the alkaloid-rich fraction (72.1 mg). The alkaloid-rich fractions were prepared as 5 mg/mL in 50% aqueous MeOH for UHPLC–ESI-qTOF-MS analysis.
All experiments were performed with a Waters Acquity UPLC system (Waters Co., Milford, MA, USA) hyphenated with a Waters Xevo F2 qTOF system (Waters MS Technologies, Manchester, UK), which was equipped with an Acquity BEH C18 column (100 mm × 2.1 mm, 1.7 μm). The mobile phase consisted of A (10 mM ammonium formate in H2O) and B (acetonitrile), with a linear gradient of 10% to 65% B (0.0-20.0 min), followed by 5.0 min of washing with 95% B and 10.0 min of isocratic 10% B for re-equilibrating the column. The flow rate of the mobile phase was set to 0.3 mL/min, and the column temperature was maintained as 25℃. Each sample (2.0 μL injected in the partial loop in needle-overfill mode) was analyzed in positive ion mode in the 100-1500 Da range with acquisition times of 0.3 s in the centroid mode. The ESI conditions were set as follows: capillary voltage 2500 V, cone voltage 45 V, source temperature 120℃, desolvation temperature 300 ℃, cone gas flow 50 L/h, and desolvation gas flow 800 L/h. The low and high collision energy for the MSE experiments were 0 eV and 20 to 35 eV, respectively.
Initially, the analytical conditions were optimized. Considering the basicity of cyclopeptide alkaloid molecules, several pH-modifying additives were added to the mobile phase. The use of a MS detector limited the use of many well-known pH modifiers, but ammonium formate was found to enhance the separative resolution of the UHPLC system. Both the positive and negative ion modes were tested, and the positive ion mode was selected because it showed a better ion intensity for cyclopeptide alkaloids.
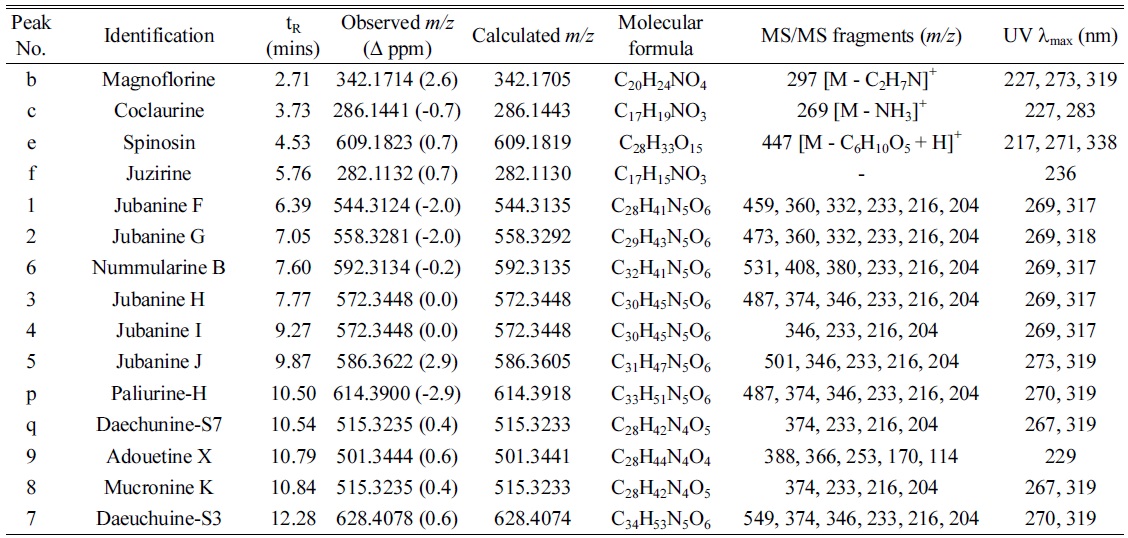
Under the optimized conditions, extracts of Z. jujuba roots and Z. jujuba var. spinosa seeds were analyzed. Because of the low contents of cyclopeptide alkaloids (0.0002% to 1% according to the reference2), they were barely detected when the total extracts were analyzed. Therefore, the alkaloid-rich fractions were prepared for the further analyses. The UHPLC–ESI-qTOF-MS chromatograms obtained for the alkaloid-rich fractions of Z. jujuba roots and Z. jujuba var. spinosa seeds are shown in Figure 1. A mixture of compounds 1-9 was injected for peak-picking, and these were identified in the chromatograms of the alkaloid-rich fractions by comparing the retention times and MS spectra. As shown in the figure, most of the isolated compounds were major constituents of both fractions, with the exception of 4 and 5, which were barely observed in Z. jujuba var. spinosa seeds. Other unidentified major nitrogen-containing chromatographic peaks were annotated as a to y, and their MS, MS/MS, and UV absorption spectra were analyzed (Table 1). Peaks a-f were not cyclopeptide alkaloids, but, with the exception of e, were smaller alkaloid molecules. The largest peak b was assumed to be magnoflorine from its molecular ion at m/z 342.1714 [M]+ (calcd. for C20H24NO4, 342.1705), which is known as one of the major components of Z. jujuba var. spinosa seeds.16 Similarly, peak e was suggested to be spinosin, the most abundant flavonoid glycoside of the plant (m/z 609.1823 [M + H]+, calcd. for C28H33O15, 609.1819). To confirm the chemical structures of these compounds, reference standards of magnoflorine and spinosin were injected using the identical analytical method, and their identifications were confirmed as described above by comparing the retention times and MS spectra. Peaks c and f were putatively identified as coclaurine and juzirine from their pseudomolecular peaks m/z 286.1441 [M + H]+ (calcd. for C17H20NO3, 286.1443) and 282.1132 [M + H]+ (calcd. for C17H16NO3, 282.1130), respectively, along with chemotaxonomic considerations.17
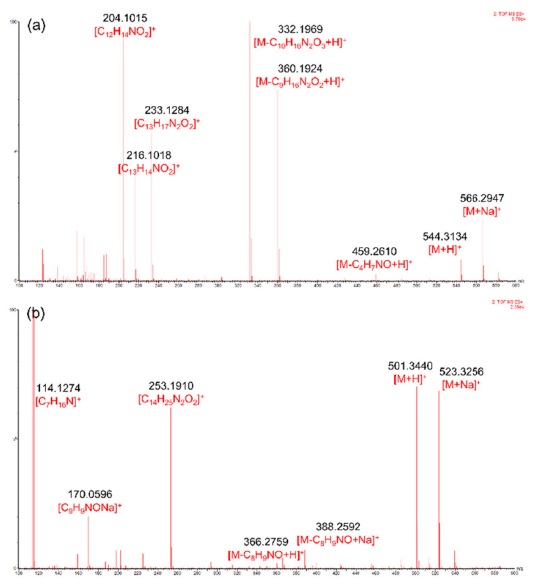
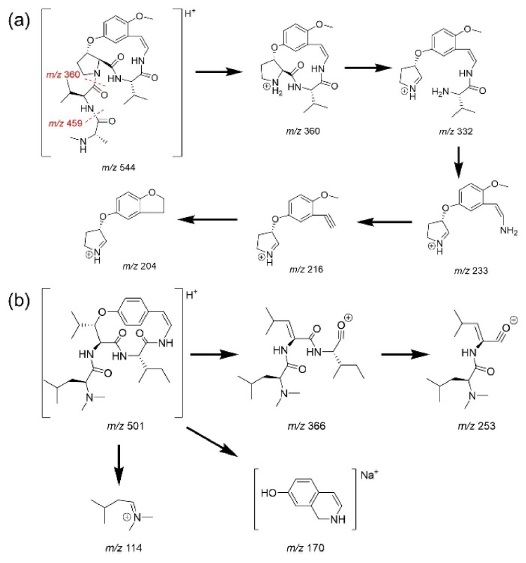
Tandem MS fragmentation pattern analysis is a very useful tool for structural determination of small and large peptides because of their polymeric structures.18 However, the fragmentation behavior of cyclopeptide alkaloids has rarely been studied; the exception being the study by Shah et al., who investigated the fragmentation mechanism of N-formylmauritine-C and N-formylsativanine-C, type-Ia and -II cyclopeptide alkaloids, respectively.19 Fragmentation reactions under the ESI process are different from those in EI-MS, and hence the MSE spectra of the identified cyclopeptide alkaloids were examined.
The MSE spectra of compounds 1 and 9 are shown in Figure 2. Type-Ib cyclopeptide alkaloids tend to yield more diverse daughter ions compared to type-Ia. This was speculated because of the difference in the stability of the macrocyclic ring structures. Strain that occurs in the 14-membered rings of type-Ia compounds hinders UV absorption in type-Ia alkaloids despite the presence of a styrylamine unit, whereas type-Ib molecules show absorption bands at around 270 and 320 nm.2 The suggested fragmentation behaviors of compounds 1 and 9 are shown in Scheme 1-(a) and -(b), respectively.
For other identified type-Ib compounds, the fragment ions of m/z 233, 216, and 204 were also observed because they have similar backbone structures. However, other fragments such as showed a neutral loss of 82 followed by 99, which are equivalent to C4H7NO for N-methylalanine and C5H9NO for valine, respectively. Furthermore, 3 and 4 showed fragments at m/z 374, which suggests they possess leucine as ring-bound amino acids instead of valine in 1. Similarly, compound 6, which consists of phenylalanine, yields a fragment ion of m/z 408. Using this method, some type-Ib cyclopeptide alkaloid peaks could be tentatively identified. Peaks p and q were identified as paliurine-H and daechuine-S7, respectively. Tandem MS spectra of type-Ia compounds did not yield many fragment ions; therefore, little information could be gathered from them. However, adouetine-X showed that most of the generated fragments are protonated ions of the terminal amino acid side chain, and thus these could also provide a little information about the partial structures of cyclopeptide alkaloids. Chemical structures identified directly and tentatively are shown in Figure 3.
The analytical conditions optimized in this study were successfully used for holistic analysis of the alkaloid constituents of Z. jujuba var. spinosa seed extract. The MSE technique enabled us to acquire sufficient tandem MS fragmentation information to tentatively identify known and unknown cyclopeptide alkaloids in the extract. This method can be useful for systemic searches of new bioactive cyclopeptide alkaloid compounds from Ziziphus species and other Rhamnaceae plants.