The considerable time and labour requirements of existing methods of microbial identification with the gradually relevant and pressing need for rapid and reliable techniques to identify microorganisms have inspired interest in the development of alternative microbial identification approaches.1 Polymerase chain reaction (PCR) of microorganisms remains the gold standard and although reliable, it is more expensive and requires a higher degree of expertise.1,2
Mass spectrometry-enabled methods have been the subject of intense interest over the past decades.3-10 The wide and rapid applicability of mass spectrometry have inspired optimism that this technology has unique potential to revolutionize microbial identification. MALDI TOF MSbased approaches have been one of the most popular expertise used in this field, first due to the relatively simple sample preparation required and secondly because of the short analysis time, high sensitivity accuracy and the speed at which data can be acquired and analysed.11,12 Straintyping can allow for discrimination of pathogenic from non-pathogenic strains and drug resistant from drug susceptible bacteria.1 A recent review demonstrated that although identification of bacteria to the strain level has been achieved after 17 years of development, several difficulties with the conventional method still pose a major challenge for wider application.1 We believe that the utility of this technology at strain-typing level requires a paradigm shift from conventional methods. We propose expansion and optimization of a recent approach that has the potential to realize the aspiration to routinely discriminate between bacteria at the strain level. This will require more flexible software control of mass spectral profiling methods by focusing on unique matchless low intensity peaks.
Studies focusing on the protein MS profiling to identify antimicrobial resistance have also been published, such as for methicillin resistant
The aim of this study was to explore the potential of mass spectral protein profiling for the identification of
>
Bacterial isolates and preparation of intact cell
Bacterial isolates of
Mass spectra were acquired using a MALDI-TOF mass spectrometer (Bruker Daltonics) equipped with the nitrogen laser. Spectra were registered in the linear mode of positive ions (20 kV) in the range of molecular masses m/z 2000-20000. The spectra were externally calibrated using the Bruker Bacterial Test Standard (BTS) obtained from Bruker Daltonics Germany. Each spectrum was acquired with 240 laser impulses (40 impulses with various spot points). The programmed software (flexControl version 3.4 build 119, flexAnalysis version 3.4 build 70 and ClinProTools version 2.2 build 83) was employed for acquiring, treatment, and analysis of mass spectra for differentiation of isolates at the strain level to differentiate drug resistant strains from the susceptible ones investigated. This clustering version of mass spectra is an approach based on the use of algorithms of the ClinProTools software, which was initially developed for search of protein markers of various oncological diseases.16,17 Identification was performed using the MALDI Biotyper 3.1 software (Bruker Daltonics) by comparing experimental spectra with the reference library of the database containing 4613 spectra of various microorganisms. Species identification was considered reliable, when the score calculated exceeded 2.300. The score from 2.000 to 2.299 provided reliable genus identification only.
>
Optimisation of the method with carbapenemase-producing E. coli
MALDI-Biotyper identification with standard methods
Primary bacteriological species identification of all selected
>
ClinProTools analysis of spectra
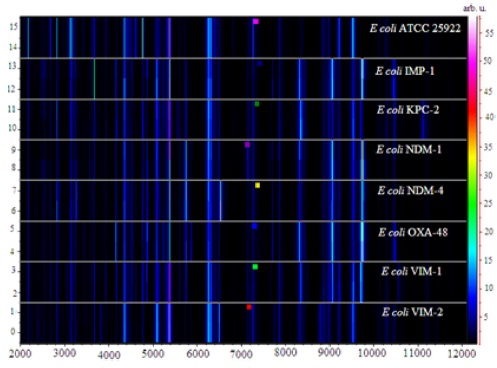
A virtual gel view representation of data derived from
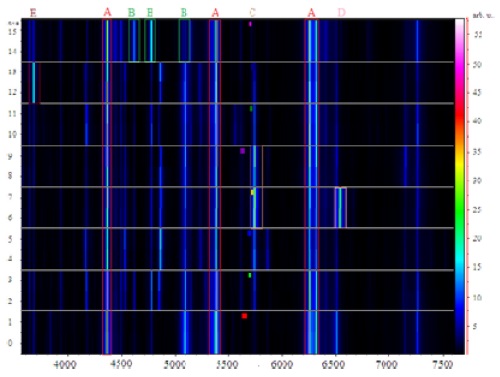
The common peaks generated by these isolates appearing at m/z 4364, 5383, and 6256 were used for normalization of the raw spectra to provide a gel view (Figure 1b). We selected a region that incorporated the common peaks to illustrate the effectiveness of visual peak picking to assist in differentiating between the isolates. The drug susceptible
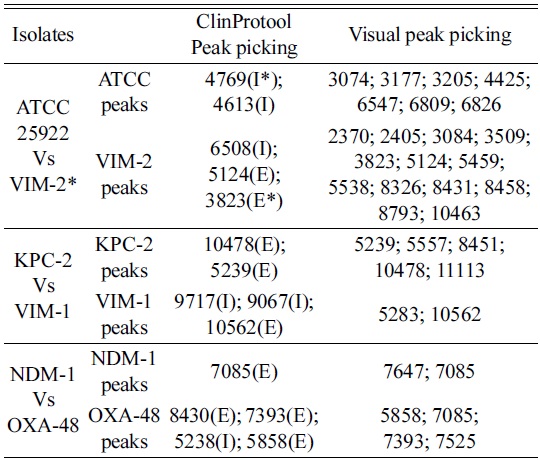
Three groups of isolates were chosen randomly to investigate the significance of using this biomarker approach to identify several mass peaks that could result in a fingerprint. ClinProTools provides a list of peaks selected according to statistical importance based on the intensity and/or the presence of peaks only, to differentiate between the selected isolates. The visual peak picking was based on the presence of identifying exclusive peaks between spectra, and this enabled the detection of significant differences amongst
[Table 1.] Comparison of ClinProTools statistics vs visual peak picking randomly
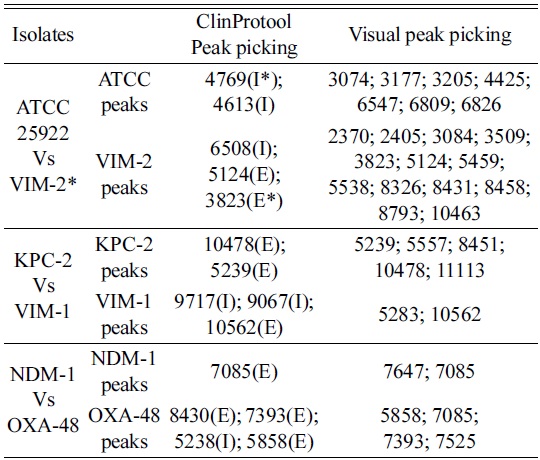
Comparison of ClinProTools statistics vs visual peak picking randomly
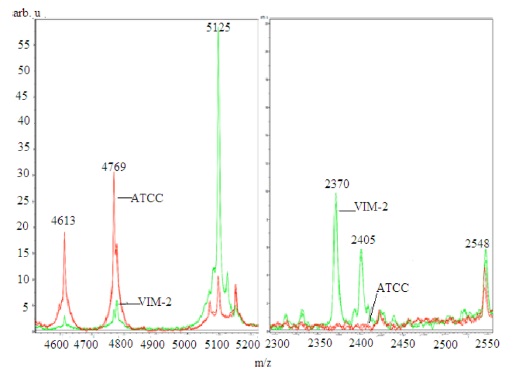
In order to differentiate the selected isolates, the ClinProTools specified a list of peaks arranged according to the statistical significance where the notified distinction was the changes of intensity in general, Figure 2 (left). Moreover the visual investigation was based only on the exclusive unique peaks Figure 2 (right). The default statistical peak lists did not provide enough significance to the lower intensity exclusive peaks as compared to intensity differences between common peaks. Current software does not allow manual changes to the default method.
>
MALDI-Biotyper identification with modified MSP methods
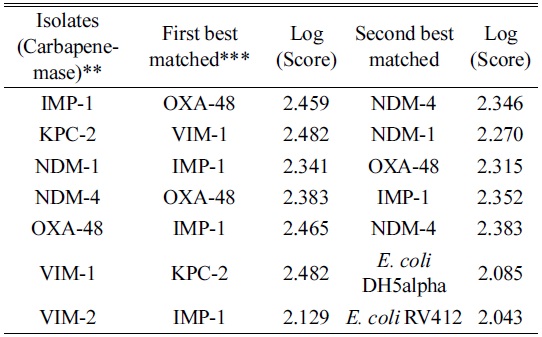
We generated a mass spectrum profile (MSP) of the standard control strain ensuring that the creation method included all of the peaks that were identified as exclusive between the species. MSP is a reference spectrum or more accurately a reference peak list that is assigned to a species or strains. Subtyping MSPs are used to distinguish between closely related species. In addition to extracting information on peak frequency and peak intensity distribution, subtyping MSPs apply additional weighting to distinguish unique peaks. The desired mass error for the MSP creation method was changed from 250 ppm to 2000 ppm and the desired peak number from 70 to 100. The mass error of the raw spectra for the identification method was restricted to be 250 ppm as compared to the standard of 2000 ppm. The modifications of the mass error were based on maximizing the inclusion of all relevant peaks (including low intensity peaks). VIM-2 was scanned against the existing Biotyper database to compare the standard methods with the modified version and, with the inclusion of the newly created MSP of the
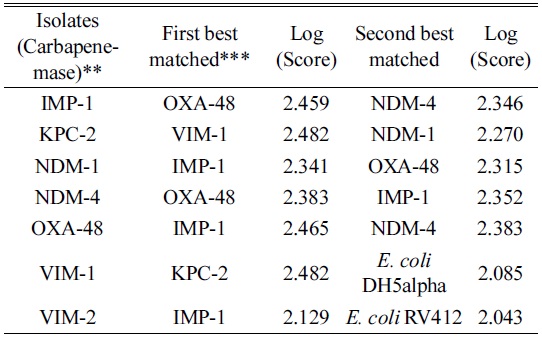
Evaluation of carbapenemase-producing E. coli isolates by the modified MSP* creation and identification methods
We scanned the CRE producing
>
Application of the optimised method to differentiate CRE strains
The second part of this study involved an application of this strategy for potential identification of different strains of carbapenem resistant bacteria.
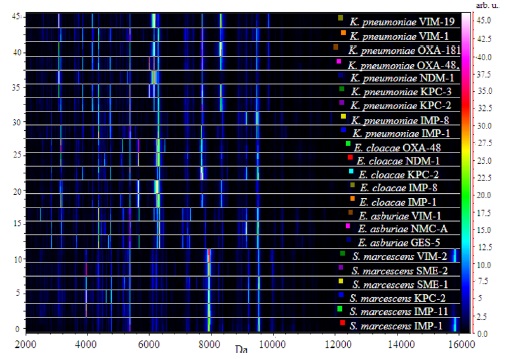
The gel view representation of the acquired spectra from
Within carbapenemase-producing
Specific bands could also be observed in the group of
In the group of
Thus, based on our data, MALDI Biotyper has potential as a tool for rapid detection and diagnosis of carbapenemase-producing bacteria. This potential can be further enhanced if the software would allow for manual weighting of the desired biomarker peaks. Alternatively, specific protein targets (biomarkers) can be used. The differentiation of carbapenemase-producing