Microalgae research is gaining momentum because of their potential biotechnological applications, including the generation of biofuels. Genome sequencing analysis of two model microalgal species, polar free-living Coccomyxa sp. C-169 and symbiotic Chlorella sp. NC64A, revealed insights into the factors responsible for their lifestyle and unravelled biotechnologically valuable proteins. However, genome sequence analysis under-explored cytochrome P450 monooxygenases (P450s), heme-thiolate proteins ubiquitously present in species belonging to different biological kingdoms. In this study we performed genome data-mining, annotation and comparative analysis of P450s in these two model algal species. Sixty-nine P450s were found in two algal species. Coccomyxa sp. showed 40 P450s and Chlorella sp. showed 29 P450s in their genome. Sixty-eight P450s (>100 amino acid in length) were grouped into 32 P450 families and 46 P450 subfamilies. Among the P450 families, 27 P450 families were novel and not found in other biological kingdoms. The new P450 families are CYP745-CYP747, CYP845-CYP863, and CYP904-CYP908. Five P450 families, CYP51, CYP97, CYP710, CYP745, and CYP746, were commonly found between two algal species and 16 and 11 P450 families were unique to Coccomyxa sp. and Chlorella sp. Synteny analysis and gene-structure analysis revealed P450 duplications in both species. Functional analysis based on homolog P450s suggested that CYP51 and CYP710 family members are involved in membrane ergosterol biosynthesis. CYP55 and CYP97 family members are involved in nitric oxide reduction and biosynthesis of carotenoids. This is the first report on comparative analysis of P450s in the microalgal species Coccomyxa sp. C-169 and Chlorella sp. NC64A.
Green algae belonging to the phylum Chlorophyta consist of microorganisms adapted to diverse ecological niches. Green algae are photosynthetic in nature and it is estimated that more than one billion years ago terrestrial plants emerged from these organisms (Heckman et al. 2001). These organisms played a key role during the evolutionary history of earth and became major players in global energy / biomass production and biogeochemical recycling (Grossman 2005, Blanc et al. 2010). Among algae, research on microalgae has been intensified because of their enormous biotechnological potential, including generation of biofuels (Behera et al. 2015), phytochemicals (De Jesus Raposo and De Morais 2015), cosmetics (Wang et al. 2015) and vaccines (Specht and Mayfield 2014) and their use in wastewater treatment (Tiron et al. 2015). Microalgae are also used as biomarkers for organic and inorganic pollution (Torres et al. 2008).
Two microalgal species, namely
Genome sequencing analysis of these species (Blanc et al. 2010, 2012) under-explored cytochrome P450 monooxygenases (P450s), heme-thiolate proteins present in organisms belonging to all biological kingdoms (Nelson 2013). P450s are known to involve organisms’ primary and secondary metabolic reactions. Oxidation of a wide variety of substrates in stereo- and regio-specific manner credited these enzymes’ use in the generation of drugs (Ingelman-Sundberg 2004, Guengerich 2006), fine chemicals and cosmetics (Guengerich 2002) and biofuels (Zhang et al. 2011) and as biosensors (Paternolli et al. 2004) and bioremediation agents (Guengerich 1995, Urlacher and Eiben 2006). A summary of P450s’ potential biotechnological applications has been discussed elsewhere (Syed and Yadav 2012).
P450s belonging to different biological kingdoms, including animals (inclusive of humans), plants, fungi, and bacteria, have been thoroughly investigated. Despite the great importance of microalgae and the potential biotechnological value of P450s, studies on microalgae P450s are scarce or not reported. In this study, we performed comparative analysis of P450s in the two model microalgal species
The genomes of
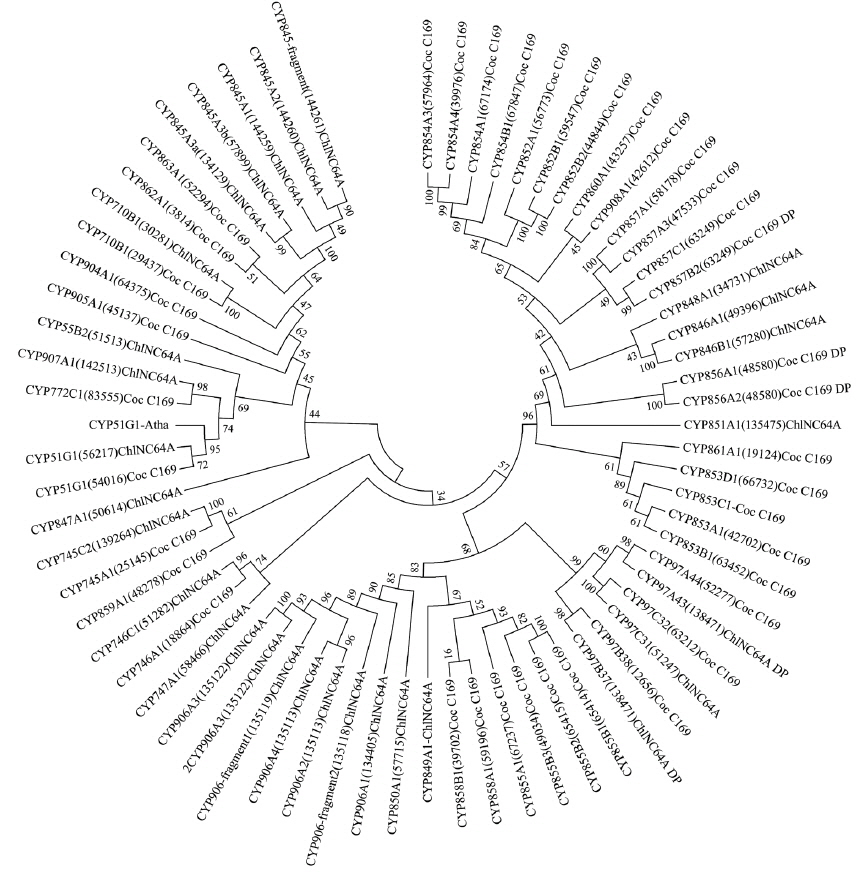
The phylogenetic relationship of the annotated algae P450s was inferred as follows: first, all the P450 protein sequences were aligned by ClustalW2 with default parameters (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Larkin et al. 2007). Then the alignment was subjected to RAxML 7.0.4 at model PROTCATDAYHOFF with 100 bootstrap replications for inferring the phylogenetic tree (Stamatakis 2006). Finally, the generated tree data were submitted to iTOL (http://itol.embl.de/) for displaying the tree (Letunic and Bork 2011).
The physical co-localization of P450 genes on the DNA was identified by searching the algal species genome database using the P450 protein ID. For each P450, its scaffold number, DNA strand and number of introns were noted and presented in tabular form. Functional analysis of algal species P450s was predicted based on their functionally characterized homolog(s) in other organisms. Furthermore, to identify the functional role, algal P450s were blasted at Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/tools/blast/) and NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins#) and their functions were predicted based on the best hit proteins with known function.
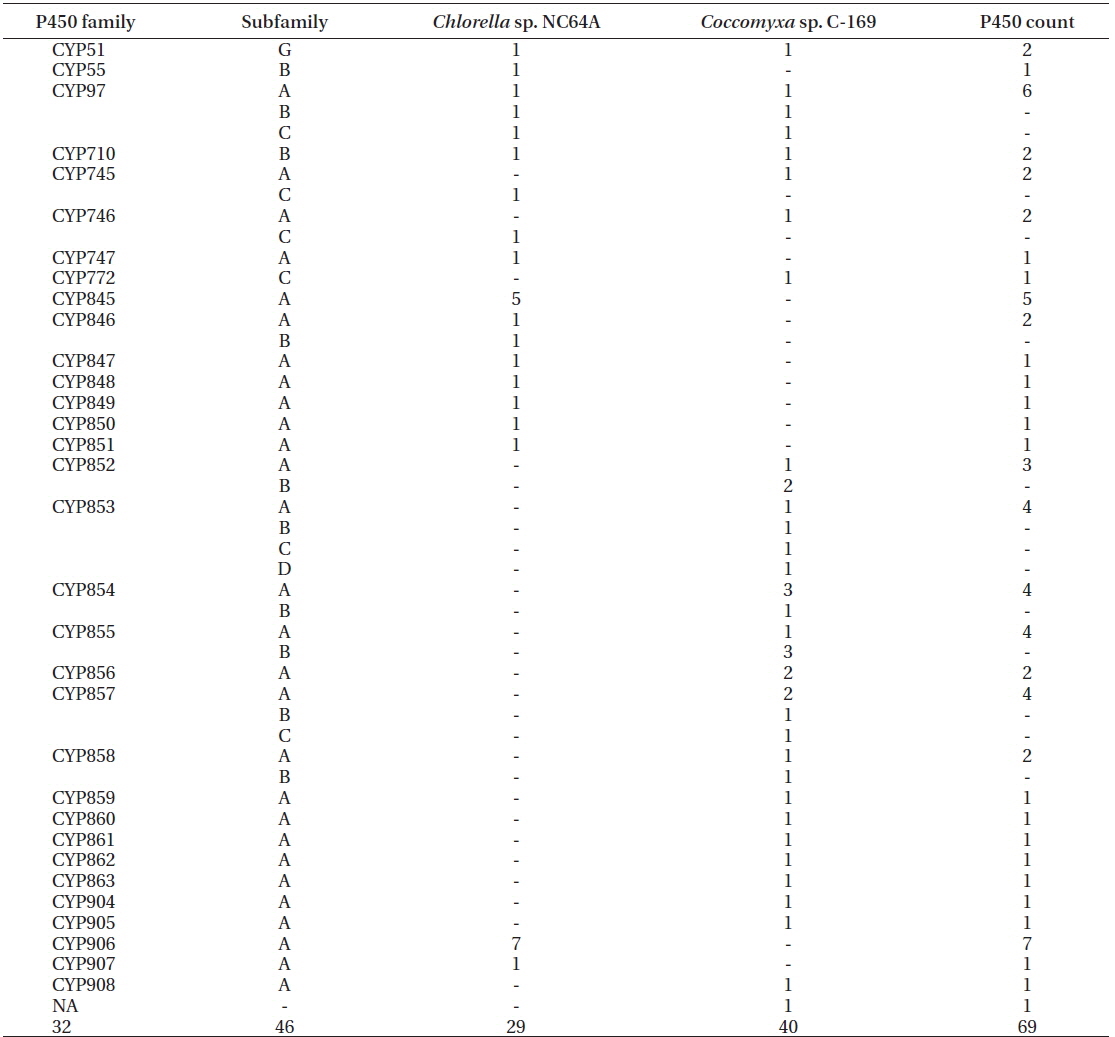
Genome-wide data mining and annotation of P450s in the two model microalgal species revealed the presence of 69 P450s (Table 1, Supplementary Fig. S1).
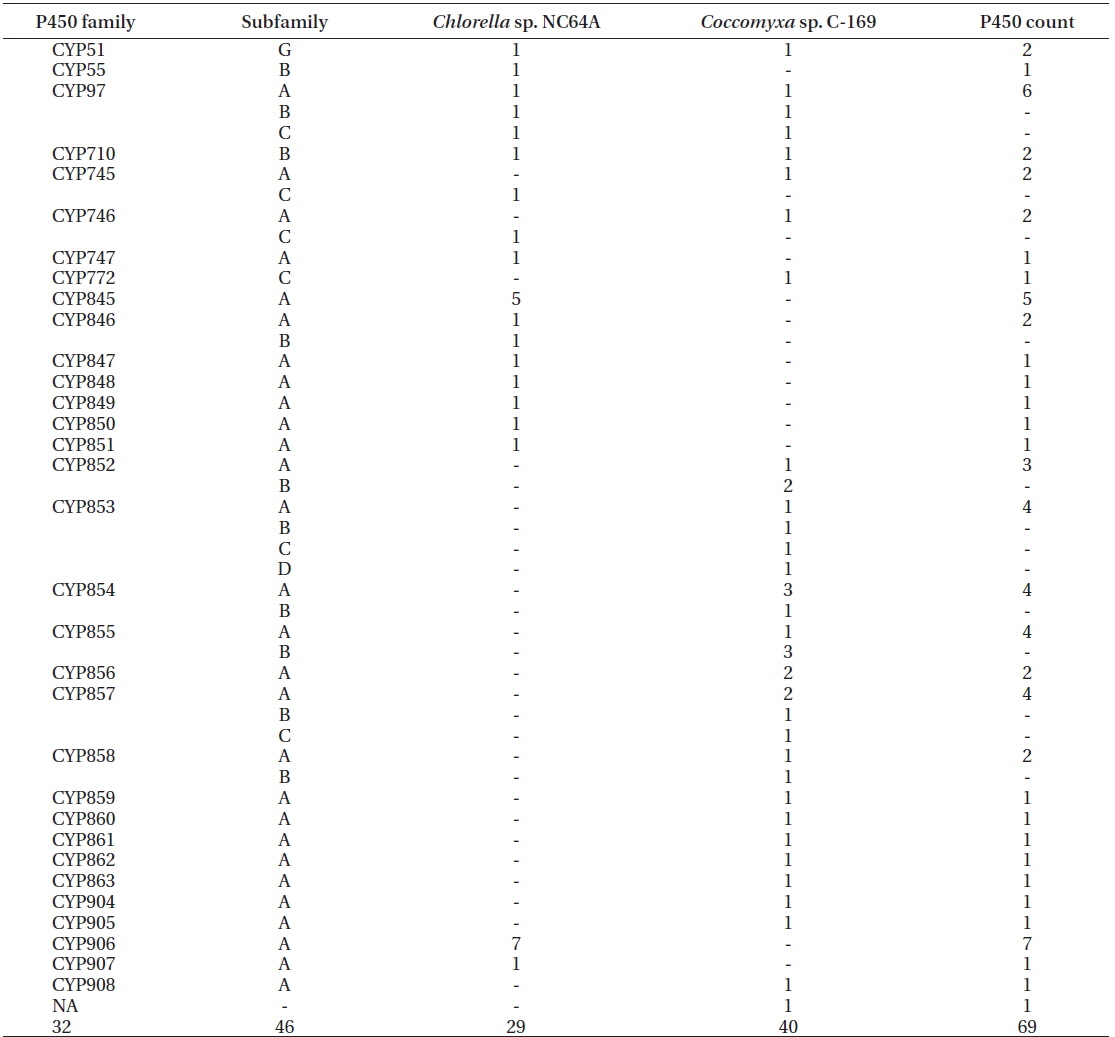
Family- and subfamily-level comparative analysis of P450s in two model microalgae, Chlorella sp. NC64A and Coccomyxa sp. C-169
The P450s of
Analysis of P450 families revealed the presence of twenty-seven new P450 families in both species. The new P450 families are CYP745-CYP747, CYP845-CYP863, and CYP904-CYP908. Comparative analysis of P450s between two algal species revealed the presence of five common P450 families (CYP51, CYP97, CYP710, CYP745, and CYP746). Sixteen (CYP772, CYP852-863, CYP904, CYP905, and CYP908) and 11 (CYP55, CYP747, CYP845-CYP851, CYP906, and CYP907) P450 families were unique to respectively
It is well known that species enrich P450s in their genome by duplication and the P450 families that contain more member P450s are termed “P450 blooms” (Feyereisen 2011). P450 blooms were observed in animals, especially in arthropods, mainly insects (Feyereisen 2011), fungi (Syed et al. 2013, 2014), and oomycetes (Sello et al. 2015) where certain P450 families bloomed because of the organisms’ need. Considering the lowest number of P450s and large number of P450 families, it is reasonable to expect no P450 blooming in the two model algal species. Interestingly, a few members of P450 families were found to be duplicated in both algal species (Supplementary Table S1). In
To date functional analysis of P450s from algal species has not reported. Hence, all algal P450s were considered orphans in terms of their function. However, based on homolog functions, P450s’ role in algal physiology can be predicted. CYP51 P450 that is conserved across the biological kingdoms is involved in the synthesis of membrane sterols (Lepesheva and Waterman 2004). The CYP97 family known as carotenoid hydroxylase is involved in carotenoid biosynthesis (Quinlan et al. 2012, Cui et al. 2013). CYP97A P450s are involved in hydroxylation of α-carotene and β-carotene at β-position, whereas CYP97C P450s are involved in hydroxylation of α-carotene at ε-position (Quinlan et al. 2012, Cui et al. 2013). The CYP97 family is highly conserved in plants (Werck-Reichhart et al. 2002, Hamberger and Bak 2013) and the presence of this P450 family in algal species suggests that divergence of this P450 family in autotrophs is preceded by the speciation of algae and plants. This suggests that CYP97 members play a key role in the generation of carotenoids and help algae in photosynthesis. Both algal species contain CYP710 P450s and studies have shown that CYP710A members are involved in sterol C-22 desaturation (Morikawa et al. 2006), the same function CYP61 has in fungi (Kelly et al. 1997), an important step in membrane ergosterol biosynthesis. The presence of CYP55, P450nor, in
Cytochrome P450 monooxygenases (P450s) play a key role in organisms’ primary and secondary metabolism. Because of their stereo- and regio-specific oxidation capability, their use in various biotechnological applications has been explored. Substantial progress has been made in understanding the animal (including human), plant, fungal and bacterial P450s in terms of their distribution, phylogenetic analysis and functional analysis. However, information on microalgae P450s is limited or not reported. This study is the first report on understanding the P450s in microalgae. Here we report identification, annotation and phylogenetic analysis of P450s in two model microalgae,