Bombyx mori has been reared as a beneficial insect in the sericulture industry and as an experimental insect in the laboratory which is highly vulnerable to viral infection (Babu et al., 2009). Bombyx mori nucleopolyhedrovirus (BmNPV) is the major pathogen causing grasserie disease. It is a member of subgroup A of Baculoviridae characterized by rod shaped enveloped virion containing a genome of circular double stranded DNA. Most of the silkworm races are susceptible to BmNPV and only few have been identified as resistant. Insect resistance to viruses has gained importance, since it plays a vital role in both the biological control of pests by viral agents and breeding of resistant strains of beneficial insects (Ponnuvel et al., 2003). The inherent immune mechanism exhibited by insects is the primary basis for their resistance to various diseases. During their evolution, insects developed a powerful and effective immune system to combat a wide variety of pathogens which had lead them to become the most diverse and successful organisms on earth. Insects posses innate immunity which is characterized by immune reactions against invading pathogens through their cellular and humoral responses. The information available on viral immune response in silkworm Bombyx mori remains scanty, except few publications on Bm lipase, Bm serine protease and few immune related proteins. (Ponnuvel et al., 2003; Nakazawa et al., 2004; Bao et al., 2009).
Infection by BmNPV is mediated by two different virions, which are occlusion-derived virus (ODV) and the budded virus (BV) (Slack and Arif, 2007). The ODV particles dissolve in the alkaline environment of the host midgut releasing ODV that fuse to the columnar epithelial cell membrane of the host intestine and initiate the infection, whereas, the BV particle consists of a single nucleocapsid, buds from the plasma membrane of an infected cell and spreads beyond the midgut through the tracheae to other parts of the host (Volkman and Goldsmith, 1985). ODV-polyhedron particles are resistant to heat and light inactivation, whereas BV is more sensitive to environment. The baculovirus envelope protein gp64 is the major phosphoglycoprotein located on the infected cells and budded virions. Insect metabolic changes play an important role in the interaction between the host and pathogen as a part of survival strategy (Babu et al., 2009).The BmNPV infection is classified into three distinct phases: early, late, and very late. The onset of late gene expression is coupled with the initiation of viral DNA replication. In the first 6 hpi , viral early genes express in host cells and are transcribed by host RNA polymerase II (Katsuma et al., 2007; Nobiron et al., 2003), whereas, late and very late genes are transcribed by an alpha-amanitin-resistant RNA, which is induced during viral replication.
The 6 hpi period is associated with BmNPV replication and both ODV and BV virion formation. (Bao et al., 2009)selected 6 hpi as a time point, to determine the resistance-related genes in the B.mori midgut, using suppressive subtractive hybridization, and found significant up regulation of eight genes from two subtracted cDNA libraries. Indian silkworm strains are known to have resistant character to viral pathogens. However, the host genes involved in immune response for resistance to invading viral pathogen have not been studied in the indigenous and exotic races. Heat shock protein genes were also found to be upregulated in most of the infected silkworm in the early stages. Based on the current understanding of the interactions between BmNPV and its host, B. mori, an attempt was made to study the differential level of gene expression at different time intervals using qPCR. The present study analyses the expression of genes conferring resistance and susceptibility to BmNPV infection in B. mori in the most divergent Indian silkworm races.
About 103 genotypes were screened using predetermined dose of the virus and from them two divergent silkworm races viz., Sarupat and CSR2 were selected as the most resistant and most susceptible to BmNPV.
B. mori multiple nucleopolyhedrovirus stock was maintained at this laboratory and used as viral inoculum. The viral inoculum was prepared by counting the number of viral polyhedra in a Neubauer chamber. The oral inoculation of BmNPV occlusion bodies was carried out in healthy newly moulted ‘0 day’ fifth instar larvae (first day after 4th moult) of Sarupat and CSR2 races with viral dosage of 40,000 polyhedral inclusion bodies (PIB) per larva.
Three replications containing twenty-five silkworms were maintained for each silkworm race. Similarly, the uninoculated control batches were reared separately under disease free environment. Silkworms feeding on BmNPV-free mulberry leaves were placed in labelled boxes until feeding was complete and then transferred to a controlled room where they remained until the end of the experiment.
BmNPV-infected fifth instar larvae were dissected, and the midguts removed at different hours post infection (hpi) (6, 12, 18, 24, 36, 48, 60 hpi). They were quickly washed in diethylpyrocarbonate (DEPC)-treated solution and immediately frozen at - 80oC for further analysis.
Ribozol RNA extraction reagents (Amresco, www.amsescoinc.com) were used to extract total RNA from the midguts of the control as well as BmNPV-infected Sarupat and CSR2 races at 6, 12, 18, 24, 36, 48 and 60 hpi. The concentration and purity of RNA was determined by measuring the absorbance at 260 nm (A260 nm) as well as the ratio between absorbance values at 260 nm and 280 nm (A260/A280), respectively. The first strand cDNA was synthesized using the RNA (2 μg/μL) treated with 1 μL each of DNase buffer and DNase (Thermo Scientific, www.thermoscientific.com) for 15 min and the reaction was terminated by heating at 65°C for 10 min. Then 1 μL of (random hexamer) primer was added and incubated at 70°C for 3 min. The incubated tubes were cooled on ice and centrifuged, 10 mM dNTP, 5X first strand buffer (4 μL), 100mM DTT (2 μL) and MMLV Superscript III reverse transcriptase (Invitrogen) 0.5 μL was added and final volume was made up to 20 μL with DEPC water which was incubated at 42°C for 60 min. The reaction was terminated by heating at 70°C for 15 min.
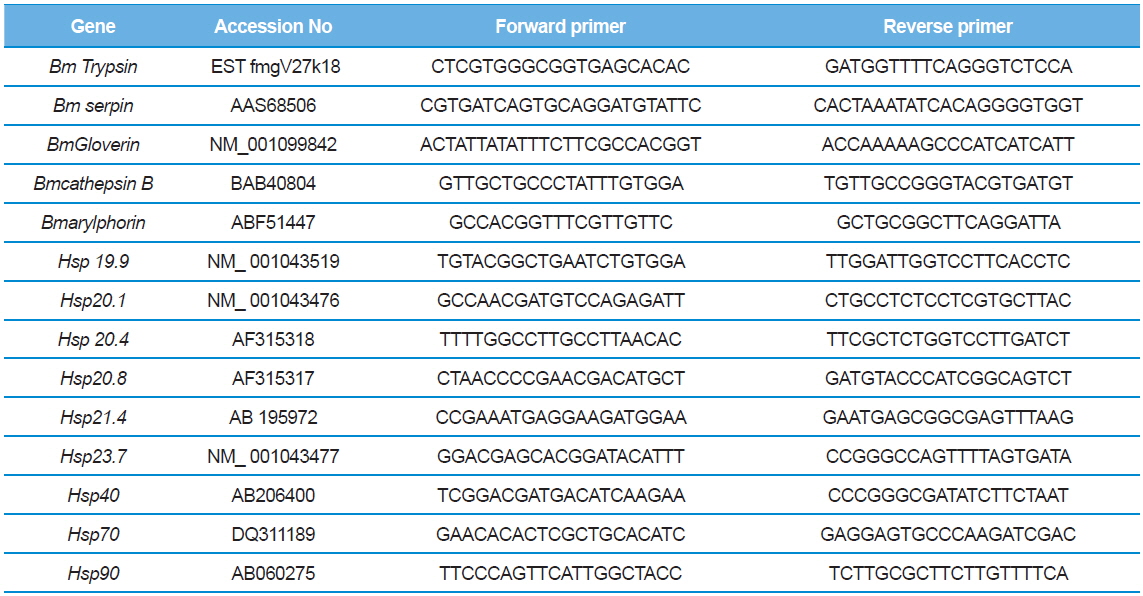
The BmNPV multiplication was estimated using qPCR with one pair of gp41 primers. The specificity of the primers and amplified products were confirmed through sequencing and subsequently analyzed using NCBI BLAST algorithm. To determine the level of gene expression associated with BmNPV resistance, which were differentially upregulated after BmNPV resistance, the qPCR experiment was performed. Five primers which were reported to be up regulated under NPV infection (Arylphorin, Lebocin, Gloverin, Serpin and Cathepsin B) and nine heat shock protein primers (Hsp 19.9, Hsp 20.1, Hsp 20.4, Hsp 20.8, Hsp 21.4, Hsp 23.7, Hsp 40, Hsp 70, Hsp90 ) were used for qPCR analysis (Table 1). One μL of first strand cDNA was used in a 20μl reaction mixture using the specific primers exclusively designed for Mx 3005P Real time PCR system (STRATAGENE). The experiment was performed in triplicate. The results were standardized to the expression level of the constitutive β actin gene. A non-template control (NTC) sample was also run to detect contamination if any.
The mortality of the races is ~39% for Sarupat (after exposure to 60000 PIB per larval dose) and 100 % for CSR2 (after exposure to 20000 PIB per larval dose) (unpublished report). The NPV doses were decided based on the LD50 value of the prototype multivoltine and bivoltine races, respectively in earlier experiments. However, in the present study, 40000 PIB/larvae was uniformly inoculated to both silkworm races and the BmNPV multiplication was monitored in the midguts of susceptible and resistant silkworm races through gp41 gene expression level. The qPCR results revealed that BmNPV invaded the midgut tissues of both the races, but multiplication rate was reduced in the resistant race from 6 hpi onwards. In order to determine the expression of genes that are activated by BmNPV infection and to understand gene expression variations related to viral proliferation, real-time PCR was performed using the larval midgut tissues of BmNPV resistant and susceptible race.
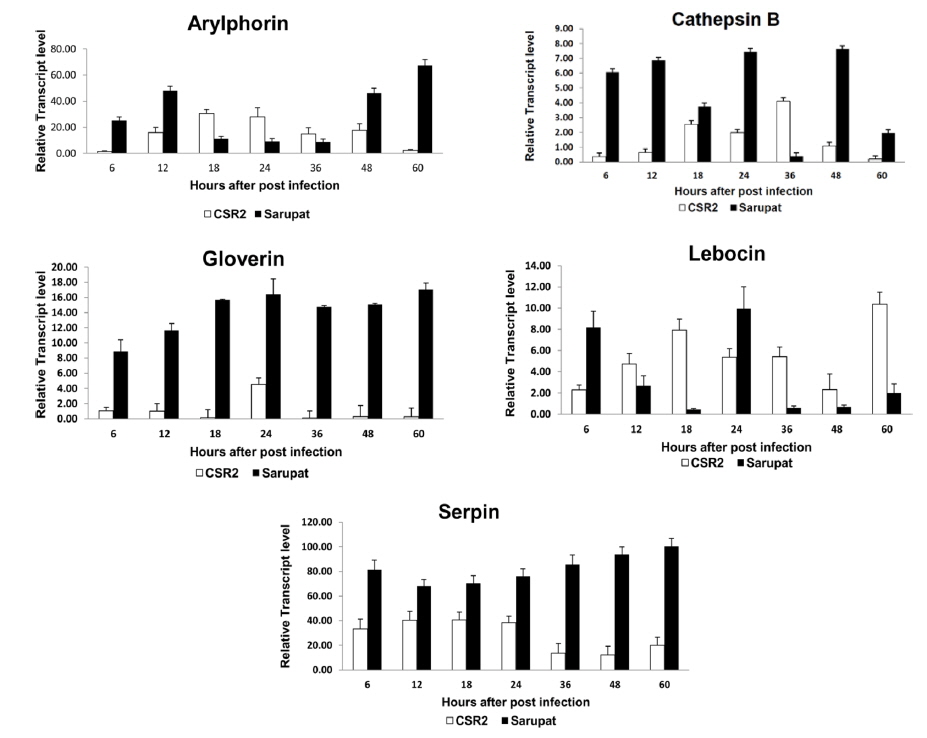
The expression of immune genes viz., arylphorin, cathepsin B, gloverin, lebocin, serpin as well as heat shock protein genes involved in the immune response in Sarupat and CSR2 races revealed differential expression levels at different time intervals (6, 12, 18, 24, 36, 48, 60 hpi) after NPV infection (Fig. 1). The transcript level of cathepsin B, gloverin, serpin, Hsp 23.7 and Hsp 40 significantly increased at all time points of post infection right from 6 hpi in the resistant race. However, the transcript level of cathepsin B was reduced at 36 hpi in resistant race. The arylphorin transcript expression level increased in resistant race until 12 hpi but significantly reduced till 36 h, followed by a quick increase at 48 hpi to reach the highest level of expression at 60 hpi. The expression level of lebocin was strongly induced in both the strains, which increased initially in resistant race with a sudden decrease in transcript level till 18 hpi, attaining maximum level of expression at 24 hpi. The transcript level in the resistant race was very low until 60 hpi in lebocin.
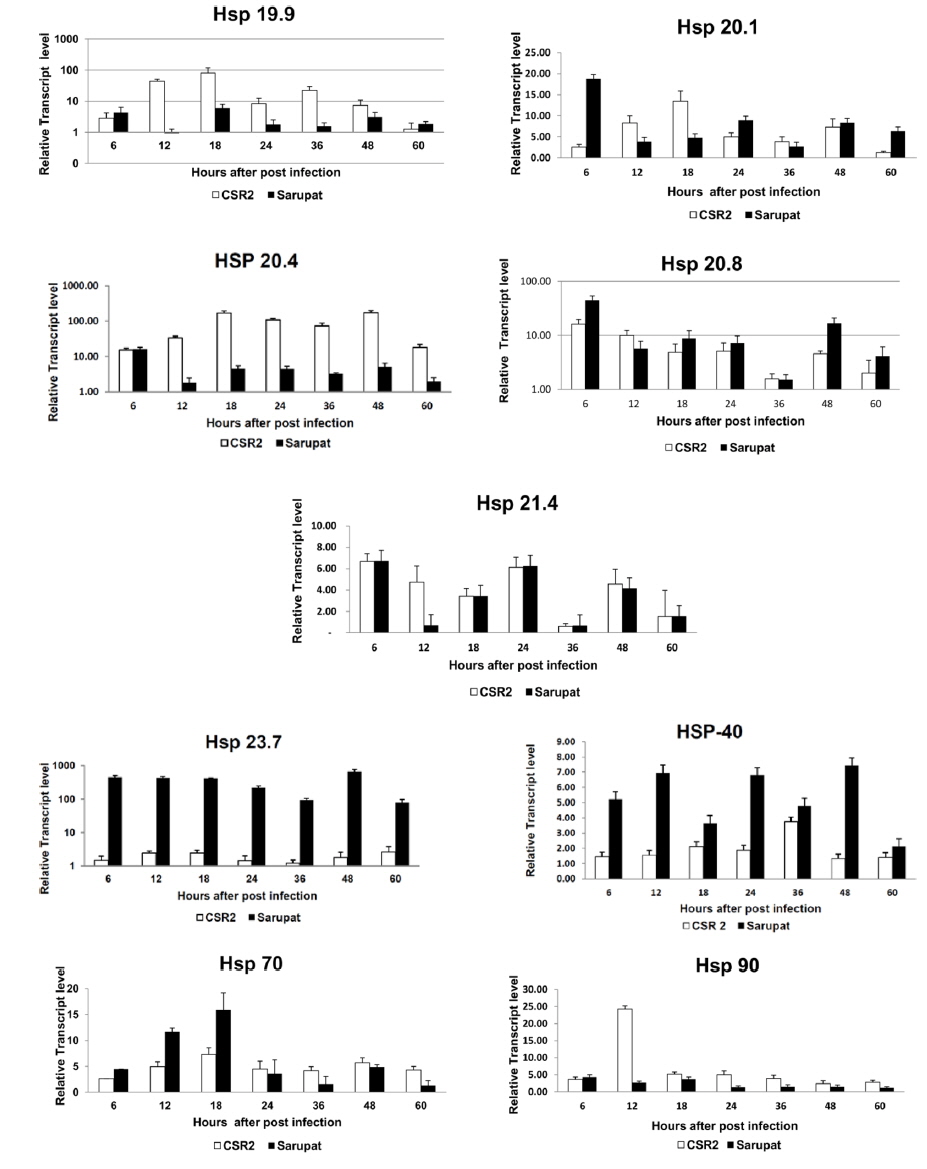
However, the transcript level of Hsp 19.9 in the resistant race increased at 6 hpi but it was very low and insignificant at 12 hpi and further the transcript level decreased in resistant race till 48 hpi of infection, followed by increase in transcript level at 60 hpi. Similarly in resistant race, Hsp 20.1 transcript level increased till 6 hpi, decreased till 18 hpi and increased again at 24, 48 and 60 hpi. In the resistant race, Hsp 20.4 and Hsp 90 transcript level increased only at 6 hpi, while, throughout the remaining time course the transcripts of both the genes consistently remained at very low levels (Fig. 2). In the resistant race, the transcript level of Hsp 20.8 showed an increase until 6 hpi that decreased at 12 hpi and then again increased at 18, 24 ,48 to 60 hpi and the maximum transcript level was at 48 hpi.
The expression level of Hsp 21.4 transcript did not show any significant change during 18 and 60 hpi in both resistant and susceptible races. Gradually, the transcript level increased a little at 24 and 36 hpi in resistant race and markedly decreased with a low level of expression at 48, and 12hpi (Fig. 2). Following BmNPV infection in resistant and susceptible races, the transcript level of Hsp 70 in resistant race gradually increased till 12 hpi to reach maximum level at 18 hpi with its transcripts constantly remaining at low level till 60 hpi in resistant races.
Although the mechanisms that underlie the regulation of immune responses in the silkworm have been studied extensively, its antiviral immune mechanism remains unclear. So far, only a few molecules that have been isolated from the B. mori larval midgut, such as lipase (Ponnuvel et al., 2003), serine protease (Nakazawa et al., 2004) and NADPH oxidoreductase (Selot et al., 2007)have been reported to exhibit antiviral activity against BmNPV infection. Many other antiviral factors may be involved in the complex immune responses of insects, but these remain undefined.
The BmNPV multiplication monitored in the midguts of highly susceptible and resistant B. mori races revealed that BmNPV invaded the midgut tissues of both B. mori races, but its multiplication was less in the resistant race. This indicated that the antiviral mechanism that occurred in the B. mori resistant race was not due to resistance against BmNPV at entry level but rather due to the inhibition of BmNPV proliferation by unknown mechanisms. Under in vivo conditions, BmNPV infects Bombyx larval midgut columnar epithelial cells within 12 hpi and mainly undergoes replication within 24 hpi (Rahman et al., 2004). Under in vitro conditions, (Katsuma et al., 2007) determined that the inhibitors of the extracellular signal-regulated kinases, ERK and JNK, markedly reduced ODV and BV production through the inhibition of the expression of delayed-early, late and very late viral genes around 12 hpi in BmNPV-infected BmN cells. Bao et al., (2009) analysed the up and down regulation of immune genes in the silkworm, B.mori using suppressive subtractive hybridization (SSH) selecting 12 hpi as a time point, since it was closely linked to viral genomic replication and virion production. These genes are implicated in multiple biological processes, but little is known with regard to their mechanisms of action in antiviral responses.
Gloverin seems to be a lepidopteron-specific antibacterial peptide (Axen et al., 1997), with expression levels that are strongly induced in Bombyx larval fat body by Escherichia coli immune challenge (Chowdhury et al., 1995; Kawaoka et al., 2008; Liu et al., 2000). In the present study, BmNPV infection caused a strong induction of B. mori gloverin gene expression in larval midguts at 24 hpi. The regulatory factors activated or blocked by BmNPV invasion, which caused a differential induction of antibacterial peptide genes in the B. mori susceptible and resistant races (Bao et al., 2009). In resistant larval midguts, these genes promptly responded to BmNPV infection at an early stage (24 hpi) indicating that they might act with each other and develop an effective defense mechanism against BmNPV infection.
The expression tendency observed in resistant larval midguts for the B. mori serpin gene was similar to that of gloverin. Serpins are serine protease inhibitors that function in regulating the prophenol oxidase activation cascade (Tong et al., 2005). B. mori serpin has a high sequence homology with an immune-responsive serpin, Manduca sexta serpin that constitutively expressed at a low level in larval hemocytes and the fat body increased dramatically upon bacterial or fungal challenge. In this study, B. mori serpin gene expression was significantly activated by BmNPV infection in resistant larval midguts at an early stage (6 hpi) to late stage (60hpi), but not activated in the susceptible race. This suggests that serpin was a quick immune-responsive gene probably involved in the host antiviral mechanism. Although insect genomes are rich in serine proteases and inhibitors, it is not well understood, how they are regulated or how microbial components trigger the activation, in any arthropod species (Xu et al., 2005). Understanding the physiological functions of these proteases and inhibitors will require further characterization to decipher their roles in immunity or other processes. Similarly, the expression patterns of the other host genes viz., arylphorin, cathepsin B, and heat shock protein genes showed differential level of expressions. The transcript levels of cathepsin B, a cysteine proteinase (Xu et al., 2001) was significantly increased in resistant larval midgut from 6 to 60 hpi, except at 36 hpi.
The heat shock protein cognate 70, a constitutively expressed member of the Hsp70 family, is the chaperone that is most commonly involved in virus interactions as well as protein folding and unfolding. It plays a role in protein translocation (Zimmermann et al., 1998) in association with several cofactors such as Hsp 40 (Hohfeld et al., 1998). Although it has not been found in association with baculovirus infection to date, it was shown to be involved in replication and assembly of other DNA viruses. Following SV40 or adenovirus type 5 infections, Hsc 70 was found to be the main virus-induced member of the Hsp70 family (Sainis et al., 1994). Hsc70 was found to interact with the L protein of hepatitis B virus, playing a crucial role in virus morphogenesis by regulating L protein translocation (Prange et al., 1999). Among heat shock proteins genes, in Hsp 20.4 transcript level was equal at 6 hpi in resistant and susceptible race and further constitutively expressed at low level in the resistant silkworm races up to 60 hpi. However, there was increase in transcript level in susceptible race from 12 hpi to 60 hpi. In summary, the antiviral mechanism that occurs in the resistant B. mori race is not due to resistance against the BmNPV invasion but rather due to the inhibition of BmNPV multiplication in the larval midgut. The defense processes against BmNPV infection that occur in the resistant larvae might be regulated via interactions involving multiple genes. The induction of heat shock proteins (HSPs) of the 70-kDa family (HSP/HSC70) in Sf-9 cells after infection with AcMNPV was monitored by Western blot analysis (Lyupina et al.,2010). They also observed that the two-dimensional electrophoresis in polyacrylamide gel revealed changes in the cellular pattern of HSP/HSC70s and synthesis of a new member of the HSP/HSC70 family in the infected cells. Although infection with AcMNPV moderately increased the HSP/HSC70 content in cells under standard conditions, the infection potentiated the response to heat shock boosting the HSP/HSC70s content in infected cells several-fold in comparison with uninfected cells. The pathways involved in these interactions are currently unclear. Our data provide information of host responses to viral infection. Further, the early up-regulation of Hsp 20.4 following NPV infection as revealed in the study, suggests that the small heat shock protein genes could play an important role in the baculovirus life-cycle. It would be an interesting protein to target for functional studies during BmNPV infection. These parental races viz., Sarupat (NPV resistant) and CSR2 (NPV susceptible) were selected through stringent procedure after considering their economic value to the sericulture industry. These races could be used for the identification of the DNA marker, developing segregating population to understand inheritance of the markers and introgression of the BmNPV resistance associated markers. Thus, these prototype potential races could contribute largely to the development of BmNPV resistant silkworm breed through marker assisted breeding program.