In the early 20th century when the first modern compilation of algal species for Bermuda was completed by Collins and Hervey (1917), only a single Centroceras species was recorded, at that time regarded as Ceramium clavulatum C. Agardh. This species was thought to be the “commonest and most variable Bermudian representative of the Ceramiaceae” (Howe 1918). Howe (1918) recognized that Centroceras clavulatum (C. Agardh) Mont. was distinguished from Ceramium by its nodal spines, and that their “length, shape, and abundance” varied greatly among individuals found in the many different habitats of the islands. Despite this, at the time Taylor (1960) completed his comprehensive flora of the warm waters of the western Atlantic, only a single species of Centroceras was again reported. Since then and prior to the advent of molecular sequencing, only two other species had been added to the western Atlantic flora, C. internitens Gallagher et Humm (Gallagher and Humm 1983) and C. minutum Yamada (Littler et al. 2008), both small epiphytes that could not be confused with the larger C. clavulatum.
Until recently, C. clavulatum was considered a poster child for cosmopolitan marine algae (Lüning 1990, Van den Hoek and Breeman 1990). It had been recorded from the Atlantic, Pacific and Indian Oceans (Guiry and Guiry 2014), and from warm temperate to tropical waters (Lüning 1990). De Barros-Barreto et al. (2006) produced an rbcL maximum-likelihood (ML) tree that first demonstrated there were at least two genetic species within C. clavulatum in the Atlantic, one from Brazil a second from the Canary Islands. A molecular-assisted alpha taxonomic (MAAT) study by Won et al. (2009) restricted the biogeographic range of C. clavulatum to the Pacific Ocean (type locality = Peru). The Won et al. (2009) study segregated out five species from the Gulf of Mexico and Caribbean, southern Chile and South Africa from a limited set of sequenced samples. They also reported that three western Atlantic species were resurrected from the ‘C. clavulatum complex;’ C. gasparrinii (Menegh.) Kütz., C. hyalacanthum Kütz., and C. micracanthum Kütz., all listed as synonyms of C. clavulatum since the middle of the 19th century (Agardh 1851). Because C. clavulatum was until now one of just two species reported from Bermuda, island collections were in need of both molecular and morphological assessment to determine their taxonomic identities, the focus of the present study.
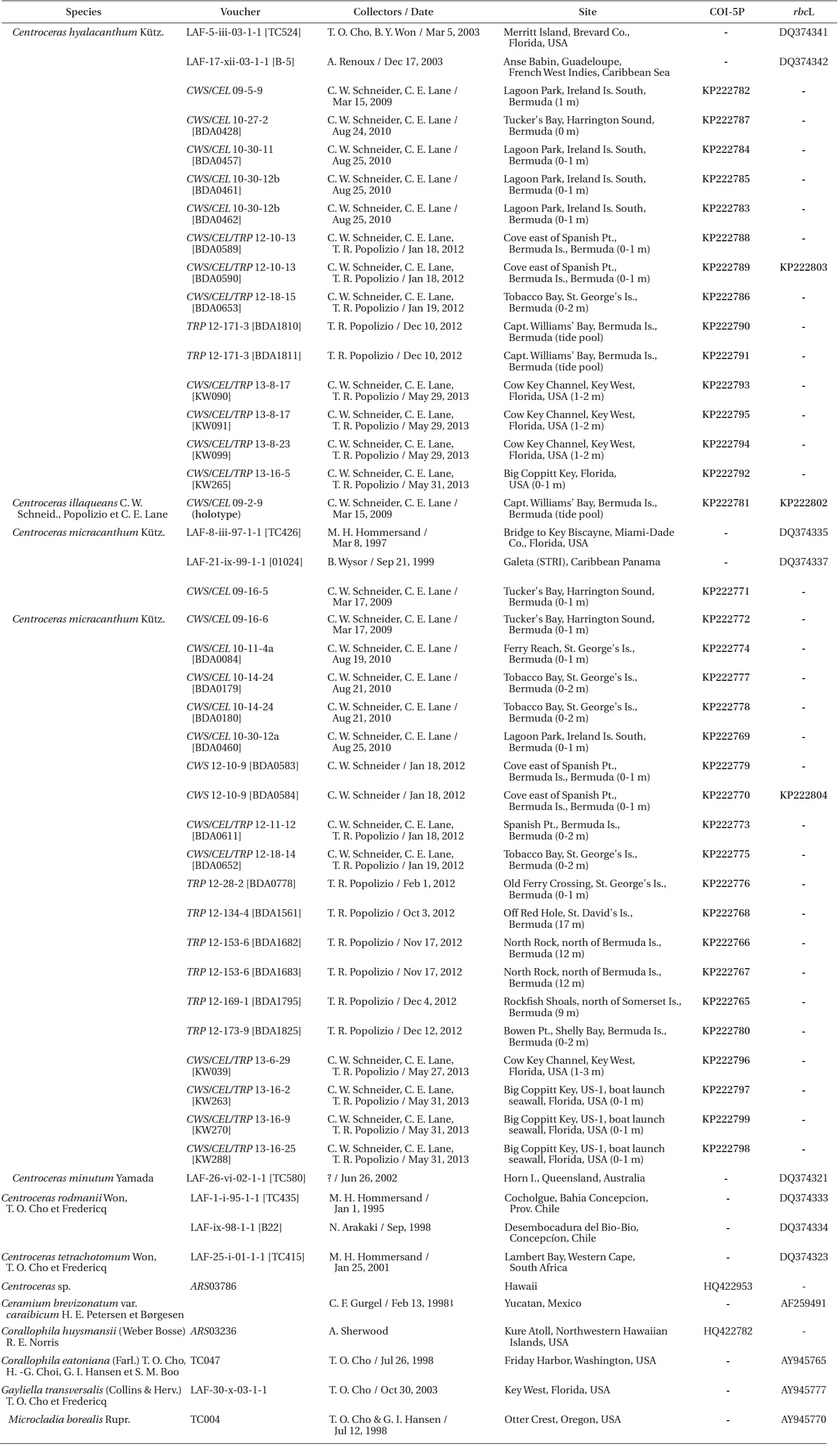
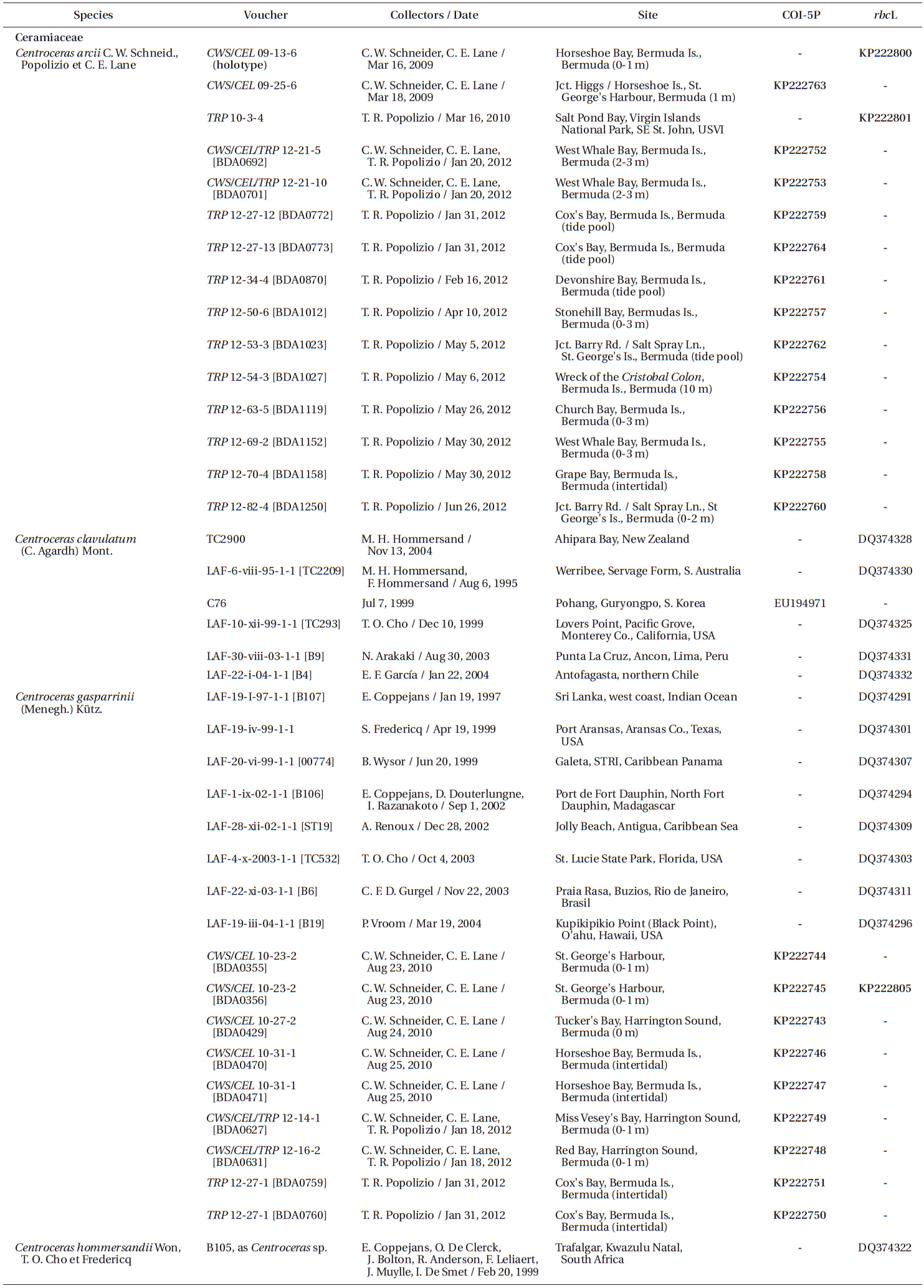
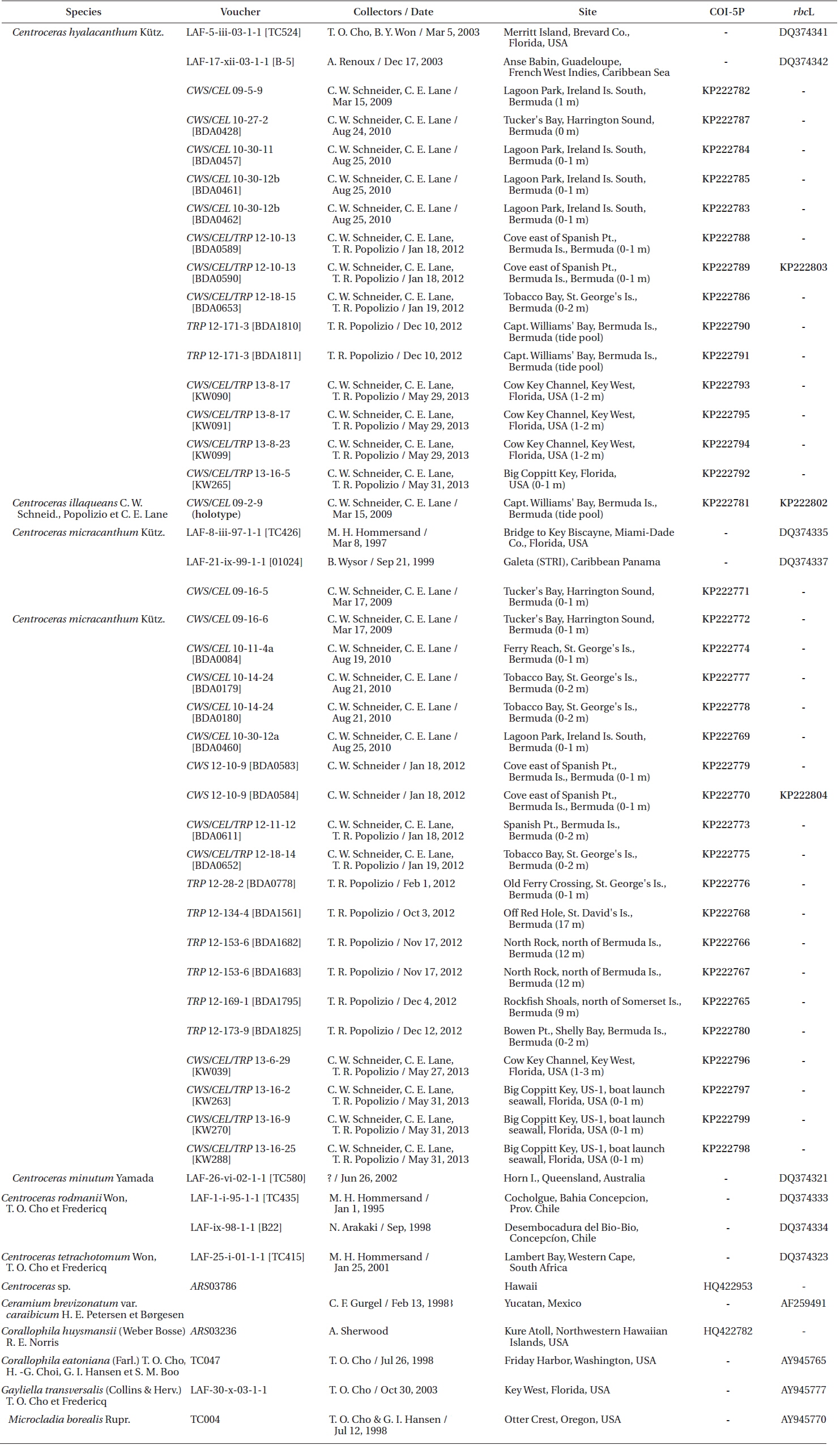
Specimens were collected by scuba or snorkeling at the depths reported in Table 1 and locations recorded using a Garmin TM GPS III Plus (Garmin Ltd., Olathe, KS, USA). All of the species were found in similar lower intertidal to shallow subtidal habitats on coral reefs or as epiphytes on other macroalgae. Only one species, Centroceras micracanthum, was found extending into deep water habitats. Portions of each specimen were dried on silica gel for DNA extraction, fixed in 4-5% formalin in seawater, and the remaining pressed as herbarium vouchers. Prior to drying, all DNA vouchers were photographed using a Canon Powershot s90 camera (Canon Inc., Tokyo, Japan). Axes with apical regions were whole-mounted onto glass slides, and liquid-preserved specimens were sectioned at 25 µm using an American Optical freezing microtome (American Optical, Buffalo, NY, USA). All of the mounts were affixed permanently in a 20 : 1 solution of 30% Karo corn syrup and 1% aniline blue. Herbarium specimens were scanned on an HP Photosmart Premium (HewlettPackard Company, Palo Alto, CA, USA), and photomicrographs were taken using Zeiss Axioskop 40 microscope (Carl Zeiss, Oberkochen, Germany) equipped with a model 11.2 Spot InSight 2 digital camera (Diagnostic Instruments, Sterling Heights, MI, USA). The digital images were assembled using Adobe Photoshop CS6 v.13.0.1 (Adobe Systems, San Jose, CA, USA). Voucher specimens are deposited in the first author’s personal herbarium (Craig W. Schneider [CWS]), with some duplicates sent to KIRI, MICH, NY and the Bermuda Natural History Museum (BAMZ). The referenced Phycotheca Boreali-Americana (P.B.-A.) exsiccata represents the set belonging to the first author, herbarium abbreviations follow the online Index Herbariorum (http://sweetgum.nybg.org/ih/), and standard author initials/abbreviations follow Brummitt and Powell (1992).
Silica-dried samples were ground in liquid nitrogen and stored at −20℃. Sample DNA was extracted from 0.5 µL ground material using the Sigma-Aldrich GenElute Plant Genomic Miniprep Kit (Sigma-Aldrich Corp., St. Louis, MO, USA) according to manufacturer protocol, with a modified lysis solution (50 µL of 10% Tween 20 and 5 µL of 20 mg mL−1 ProK), as well as 1 h at 23℃ incubation followed by a 20 min incubation on ice (Saunders and Druehl 1993). DNA was amplified via polymerase chain reaction (PCR) with the Takara Ex-Taq DNA polymerase kit (PanVera, Madison, WI, USA) in an Eppendorf AG Mastercycler epGradient thermal cycler (Eppendorf, Hamburg, Germany). To assign all specimens to species groups, two oligonucleotide primers were used for both sequencing and amplification of the COI-5P mitochondrial marker, GWSFn (Le Gall and Saunders 2010) and GWSRx (Saunders and McDevit 2012). Several specimens were selected for additional sequencing of the plastidencoded RuBisCO operon. Four oligonucleotide primers were used for both amplification and sequencing of two overlapping fragments (forward RR1 5′ ATGTCTAACTCTGTAGAAG 3′ and reverse RR4 5′ TTCAGCTCTTTCATACAT 3′; forward RrIf 5′ TCTCAGCCTTTTATGCGTTG 3′ and reverse Rrr 5′ ATCTCACTATTCTATACTCC 3′). A denaturation cycle of 94℃ for 4 min was followed by 38 cycles of 94℃ for 1 min, 50℃ for 1 min, 72℃ for 2 min, and a final extension of 72℃ for 7 min. Samples were stored at 4℃ until removed from the system. Amplified DNA was treated with Sigma-Aldrich GenElute PCR Clean-Up Kit per manufacturer protocol, and the purified PCR product was sequenced at the Rhode Island Genomics and Sequencing Center using the Applied Biosystems Inc. 3130xl Genetic Analyzer (Life Technologies, Grand Island, NY, USA).
Sequence data were aligned using the multiple sequence comparison (MUSCLE) by log-expectation) alignment program in Geneious (v.6.1.8 available from http://www.geneious.com) followed by neighbor-joining analysis of the COI-5P barcode alignment (59 specimens, 567 sites) with Tamura-nei-corrected distances (default setting). This tree was used to identify genetic species groups.
The best models of evolution for the individual gene regions COI-5P (10 taxa, 567 sites) and rbcL (30 taxa, 1,289 sites), after removing identical sequence replicates as identified in the previous analysis, were first estimated in jModelTest volume 2.1.5 (Guindon and Gascuel 2003, Darriba et al. 2012). The model GTR + I + G model was implemented for both COI-5P and rbcL. Maximum-likelihood (ML) analyses for the COI-5P data were conducted using the PHYML 3.0 (Guindon et al. 2010) plugin for Geneious, with BIONJ used to designate the starting tree, best of nearest-neighbor interchange or subtree pruning and regrafting branch-swapping options, and with the tree topology, branch lengths and substitution rates optimized. Branch support was estimated using 1,000 bootstrap replicates. Bayesian analyses were conducted using MrBayes v.3.2.2 (Ronquist and Huelsenbeck 2003) and run with four parallel chains including three heated chains, with branch lengths optimized during the run for one million generations. The initial 2,500 trees were discarded as the burn-in. Posterior probabilities were estimated based on the remaining trees. The rbcL ML phylogeny was estimated using the RAxML graphical user interface (Silvestro and Michalak 2012) with 1,000 bootstrap replicates. Bayesian posterior probabilities were generated in MrBayes, with the same parameters as in the COI-5P analysis. Stationarity was attained after the first 250,000 generations (burnin = 2,500 trees). The COI-5P (Fig. 1) and rbcL (Fig. 2) consensus trees were both manipulated for presentation using FigTree software (http://tree.bio.ed.ac.uk/software/figtree/).
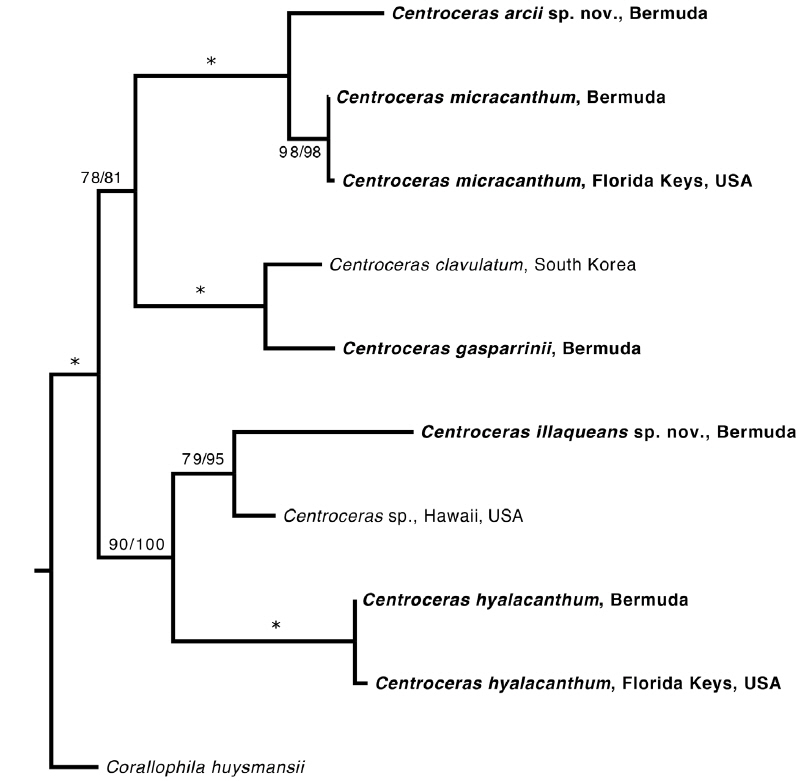
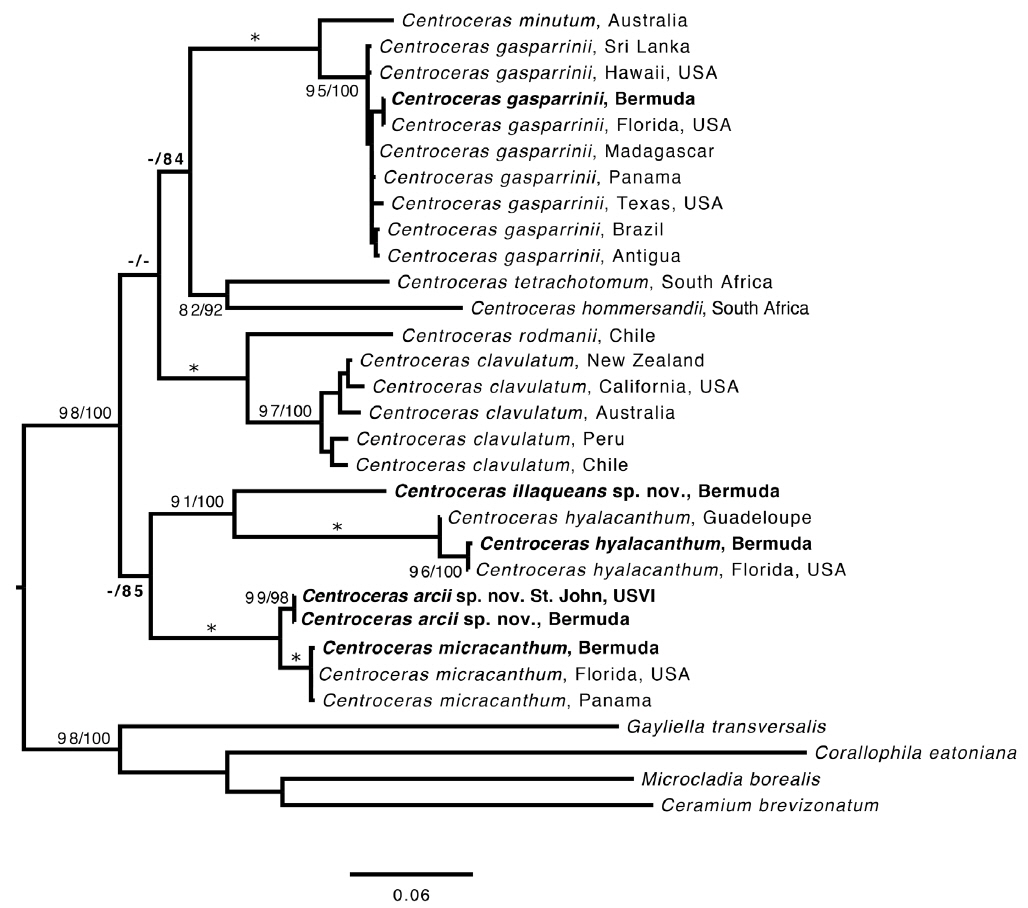
Based on our many Bermuda DNA samples and available sequences from GenBank (Table 1), COI-5P barcode analyses (Fig. 1) and rbcL phylogenies were generated (Fig. 2). As shown in these trees, the three species Won et al. (2009) resurrected for the western Atlantic Ocean by aligning type specimens with their recent genetic material, C. gasparrinii, C. hyalacanthum, and C. micracanthum, are found in the Bermuda flora. In addition, two novel species were delineated in our molecular analyses. During the 20th century, all of these five species would have been identified as C. clavulatum in the islands (Schneider 2003, p. 301). Along with the new genetic information, we follow Won et al. (2009, Fig. 14) in using nodal cortical units and the origin, number and shape of cortical cells, glands and spines produced from cortical initials as distinguishing features among the species. The descriptions presented for each species below reflect genetically determined specimens from Bermuda, Florida and the U.S. Virgin Islands.
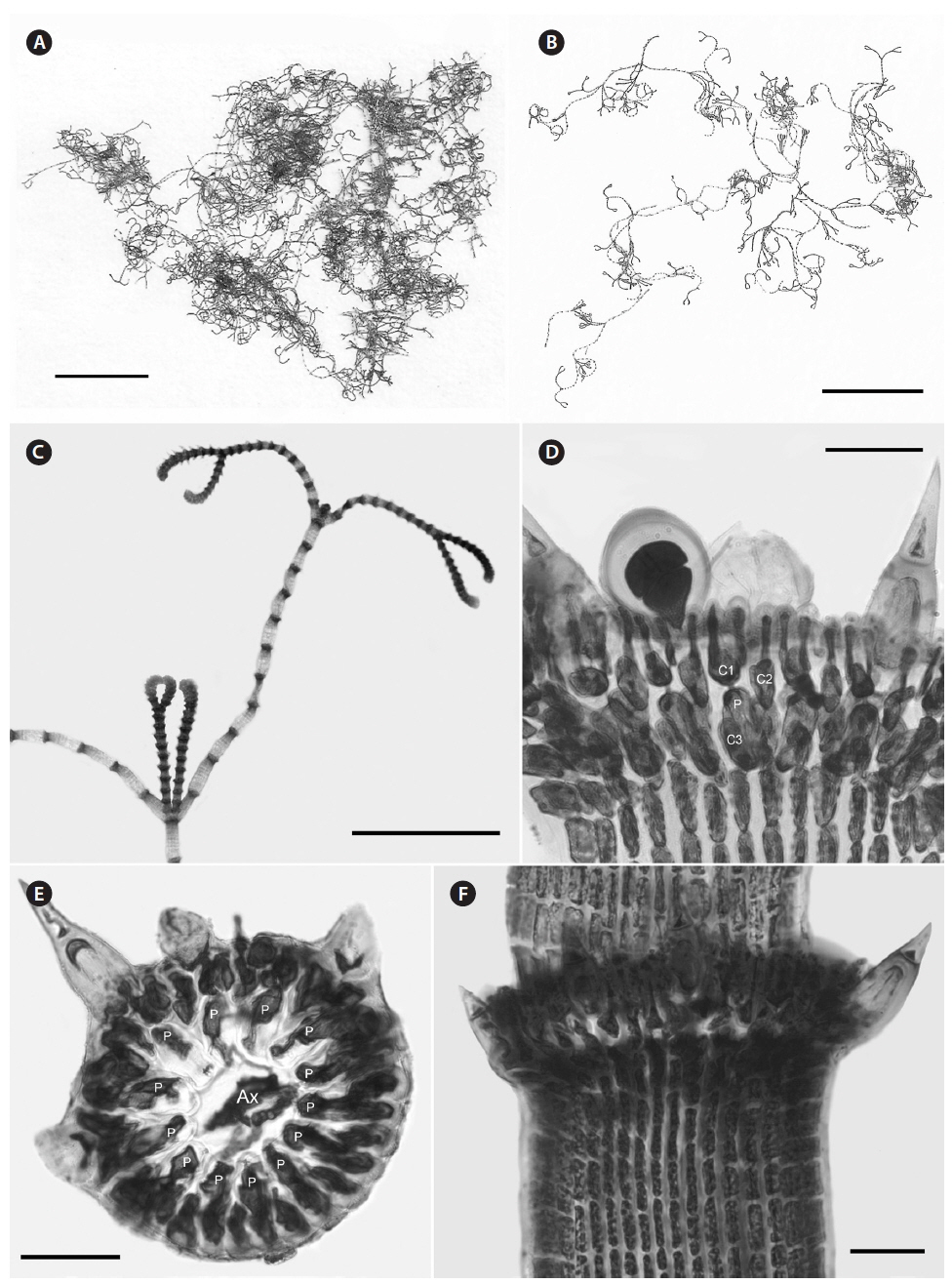
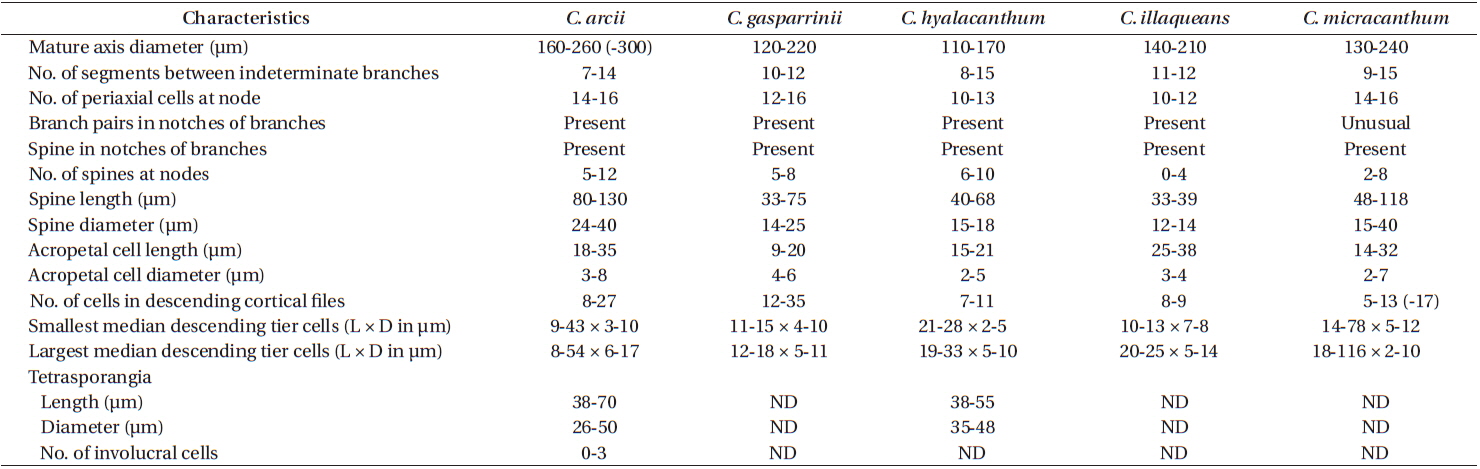
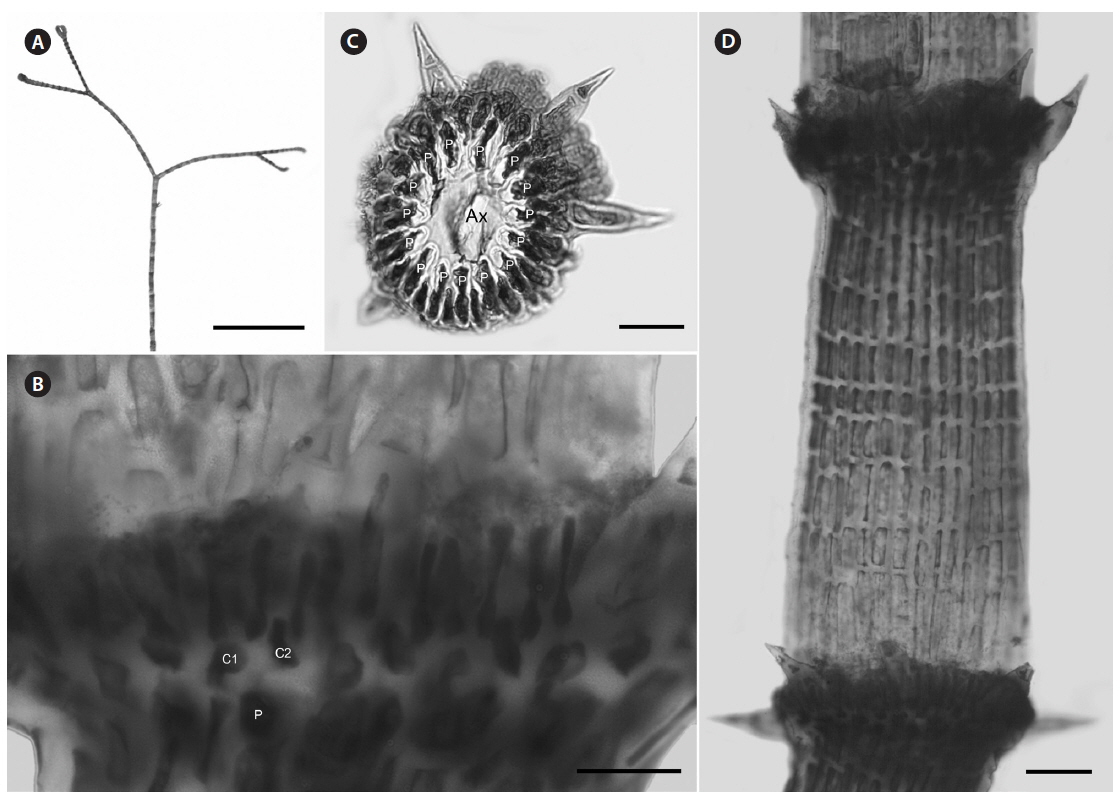
Description. Plants brownish- to rosy-red, with prostrate and erect axes 160-260 (−300) µm in diam. and to 5 cm tall (Fig. 3A & B); overall branching pattern pseudodichotomous with a branch or pair of branches usually occurring in the notches of main branches at apices (Fig. 3C); branches forming at intervals of 7-14 axial cells on the main axes, the branches divaricate and arched reflexively with straight or forcipate apices; adventitious branches developing from periaxial cells in lower portions (Fig. 3C); uniseriate, multicellular rhizoids issued from prostrate axes and lower portions of erect axes; 5-12 straight, 2-celled spines whorled at axial cell nodes, one always found in notches of dichotomous branches, 24-40 µm diam. and 80-130 µm long (Fig. 3D); distal ends of axial cells cutting off 14-16 periaxial cells, these remaining at the nodes during axial cell elongation (Fig. 3E); periaxial cells cutting off two cortical initials acropetally and one basipetally (Fig. 4A); the first cortical initial cutting off one spine and one elongate cortical cell, one gland cell and one elongate cortical cell, or two elongate cortical cells, the second cortical initial cutting off one elongate acropetal cortical cell and one basipetal cortical filament, the third cortical initial cutting off one basipetal filament; basipetal filaments made of mostly staggered cortical cell files of 8-27 cells, the cells rectangular in surface view, the smallest being 3-10 µm diam. and 9-43 µm long, the largest being 6-17 µm diam. and 8-54 µm long, these files completely corticating axial cells from node to node (Fig. 3F); tetrasporangia formed in whorls at the nodes (Fig. 3D), one produced from each periaxial cell, spherical to subspherical, 26-55 µm diam. and 38-75 µm long, subtended by 0-3 involucral filaments; gametangia not seen.
[Fig. 3.] Centroceras arcii sp. nov. (A) Habit of one of two cards in holotype packet, CWS/CEL 09-13-6. (B) Habit displaying reflexive branching, CWS/CEL/TRP 12-21-10. (C) Axis with a branch pair in the notch of the main branch and single branch in penultimate notch, CWS/CEL/TRP 12-21-10. (D) Axial node with cortical initials bearing elongated acropetal cells, spines, and tetrasporangium, CWS/CEL 08-9-2. (E) Cross section of axis showing 13 periaxial cells, TRP 12-34-4. (F) Axial segment displaying staggered, basipetal filaments of cortical cells, TRP 12-70-4. Ax, axial cell; C1-3, sequence of cortical initials; CEL, Christopher E. Lane; CWS, Craig W. Schneider; P, periaxial cell. Scale bars represent: A, 1.5 cm; B, 2 cm; C, 2 mm; D-F, 50 μm.
Diagnosis. This species is morphologically cryptic with C. micracanthum, but differs genetically in its COI-5P barcode sequences (refer to Table 1 for GenBank numbers).
Etymology. Named for A. Ralph Cavaliere (ARC), Professor Emeritus at Gettysburg College, who introduced the first author to the beauty of the algae, to commemorate his retirement as the beloved Charles H. Graff Professor of Biology. ARC spent many summers in Bermuda studying and teaching the local marine flora and publishing a number of works on seaweeds including his local field guide, Sterrer and Cavaliere (1998), Bermuda’s Seashore Plants and Seaweeds. The epithet also is suggestive of arcus (L.), a bow-like curved line forming reflexively in distal portions of this species.
Holotype (designated here). Craig W. Schneider(CWS)/ Christopher E. Lane (CEL) 09-13-6, Mar 16, 2009, Horse-shoe Bay grotto, south shore Bermuda Is., 32°15′01.1″ N, 64°49′23.9″ W, Bermuda, western Atlantic Ocean [MICH] (Fig. 3A), GenBank No. KP222800; isotypes—KIRI, NY, UNB, US, Herb. CWS.
Paratypes. Bermuda—CWS/CEL 08-9-2 (⊕), CWS/CEL 09-1-3, CWS/CEL 09-25-6, CWS/CEL/TRP 12-21-10 [BDA0701], TRP 12-27-13 [BDA0773], TRP 12-34-4 [BDA0870], TRP 12-50-6 [BDA1012], TRP 12-53-3 [BDA1023], TRP 12-63-5 [BDA1119], TRP 12-69-2 [BDA1152], TRP 12-70-4 [BDA1058], TRP 12-82-4 [BDA1250]. St. John, Virgin Is.—TRP 10-3-4 (for collection details see Table 1).
Distribution. Bermuda, Virgin Islands.
Remarks. C. arcii is the most common member of the genus in Bermuda, and is sister to the morphologically cryptic and sympatric C. micracanthum in a separate clade according to COI-5P and rbcL analyses. Sequence divergence between the new species and C. micracanthum is ~1% for rbcL and ~9% for COI-5P, the latter clearly demonstrating genetic separation.
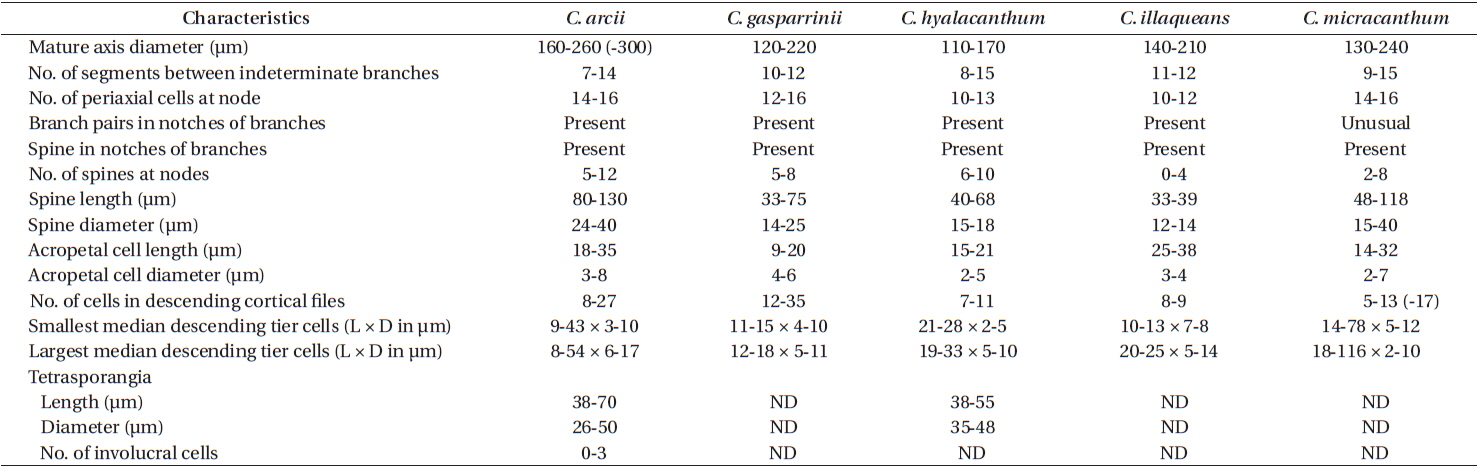
The most obvious morphological feature of C. arcii is its divaricate, reflexive branching (Fig. 3B) giving the plants an arching overall habit, a feature found in all mature collections of the species thus far. Unfortunately, C. micracanthum demonstrates at least some reflexive branching in ca. 75% of our collections and when this is the case, this species is difficult to distinguish from C. arcii. Won et al. (2009, Fig. 7b) demonstrated reflexive branching in C. micracanthum. Thus, C. arcii and C. micracanthum are cryptic in their anatomies (Table 2). At the extremes of ranges for certain characters, e.g., spine length, number of cells in descending cortical files and median sizes of tier cells, there are differences between the two species, but for the most part, they demonstrate overlapping ranges for cell dimensions and numbers (Table 2). At the extreme, cells composing basipetal cortical files are more numerous in C. arcii than those in C. micracanthum. Invariably, C. arcii has one or two branches formed in the notches of distal dichotomies, and such branches are uncommon in C. micracanthum. In the end, the two species are nearly indistinguishable and, as noted above, require gene sequencing to differentiate them with certainty.
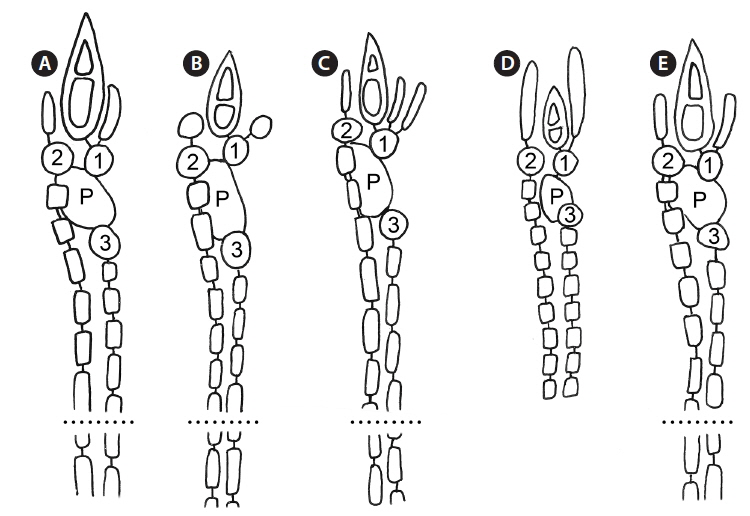
As is also true for C. micracanthum, in all but the first periaxial cell cut off in C. arcii, three cortical initials form, two acropetally and one basipetally (Fig. 4A). The first periaxial cell cuts off three cortical initials acropetally and one cortical initial basipetally for a total of four cortical initials. In all cortical initial groups, the first cortical initial cuts off one spine and a single elongate cortical cell, one gland cell and a single elongate cortical cell, or two elongate cortical cells. The production of cortical cells from acropetal cortical initials in C. arcii is the same as that found in C. micracanthum and C. illaqueans, all producing only two cells from their first cortical initials. In C. arcii, the largest spines at nodes are longer than those of any other members of the ‘C. clavulatum complex’ in Bermuda.
Basionym. Ceramium gasparrinii Meneghini 1844, p. 186.
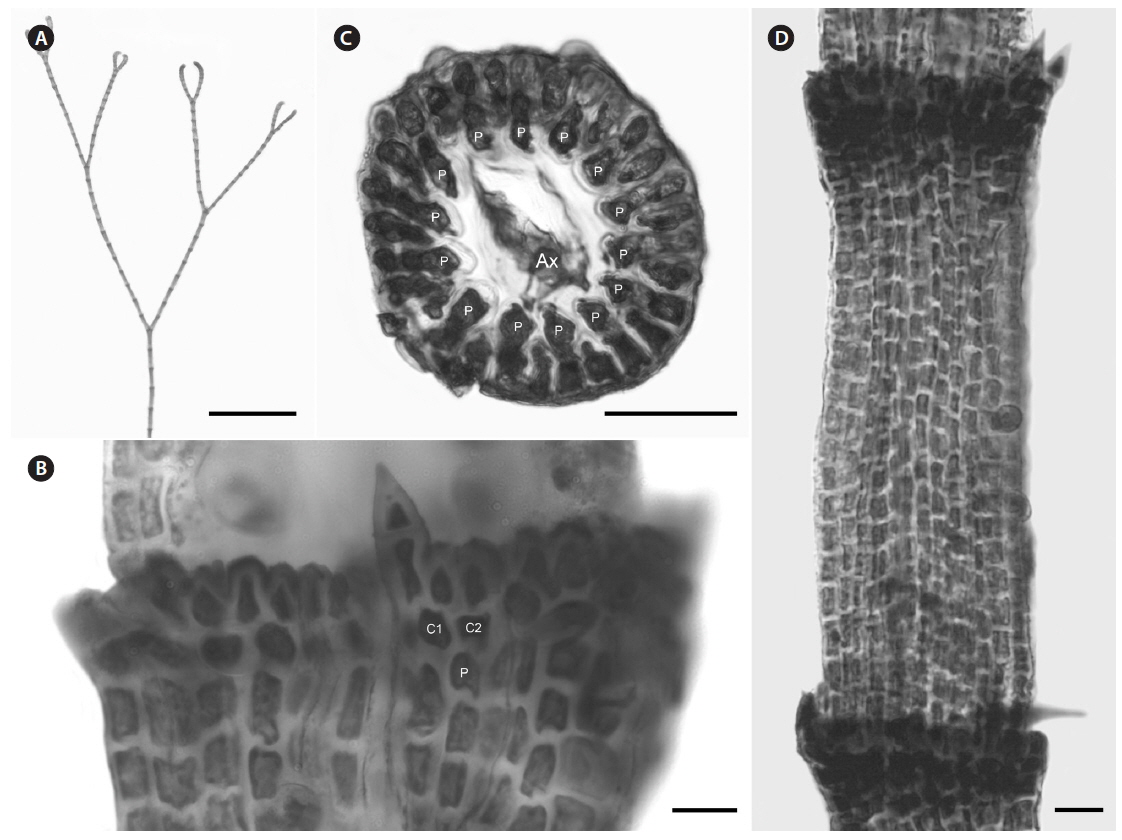
Description. Plants dark rosy-red with prostrate and erect axes 120-220 µm in diam. and to 7 cm tall; branching pattern pseudodichotomous with a branch or pairs of branches usually occurring in the notches of main branches at apices, branches forming at intervals of 10-12 axial cells on the main axes, the branches fastigiate with slightly forcipate apices (Fig. 5A); adventitious branches developing from periaxial cells in lower portions; uniseriate, multicellular rhizoids issued from prostrate axes and lower portions of erect axes; 5-8 straight, 2-celled spines whorled at axial cell nodes, one almost always found in notches of dichotomous branches, 14-25 µm diam. and 33-75 µm long (Fig. 5B & D); distal ends of axial cells cutting off 12-16 periaxial cells, these remaining at the nodes during axial cell elongation (Fig. 5C); periaxial cells cutting off two cortical initials acropetally and one basipetally (Fig. 4B); the first cortical initial cutting off one spine and one ovoid cortical cell, one gland cell and one ovoid cortical cell, or two ovoid cortical cells, the second cortical initial cutting off one acropetal cortical cell and one basipetal cortical filament, the third cortical initial cutting off one basipetal filament; basipetal filaments made up of mostly staggered cortical cell files of 12-35 cells, the cells rectangular in surface view, the smallest 4-10 µm diam. and 11-15 µm long, the largest 5-11 µm diam. and 12-18 µm long, these completely corticating axial cells from node to node (Fig. 5D); gland cells ovoid; gametangia and tetrasporangia not seen in Bermuda collections.
[Fig. 5.] Centroceras gasparrinii. (A) Axis with straight, pseudodichotomous branching, CWS/CEL/TRP 12-20-13. (B) Axial node with cortical initials bearing ovoid acropetal cells and a spine, CWS/CEL/TRP 12-14-1. (C) Cross section of axis showing 14 periaxial cells, CWS/CEL/TRP 12-14-1. (D) Axial segment displaying staggered, basipetal filaments of cortical cells, CWS/CEL/TRP 12-14-1. Ax, axial cell; C1 & 2, sequence of cortical initials; CEL, Christopher E. Lane; CWS, Craig W. Schneider; P, periaxial cell; TRP, Thea R. Popolizio. Scale bars represent: A, 1.5 mm; B, 15 μm; C, 50 μm; D, 25 μm.
Type locality. Palermo, Sicily, Mediterranean Sea.
Distribution. Widely distributed in warm temperate to tropical zones in the Atlantic, Pacific and Indian Oceans.
Selected collections. Bermuda—CWS/CEL 09-5-11, CWS/CEL 10-23-2 [BDA0355], CWS/CEL 10-27-2 [BDA0429], CWS/CEL 10-31-1 [BDA0470], CWS/CEL/TRP 12-14-1 [BDA0627], CWS/CEL/TRP 12-16-2 [BDA0631], CWS/CEL/TRP 12-20-13 [BDA0685], TRP 12-27-1 [BDA0760], TRP 12-32-5 [BDA0841] (for collection details see Table 1).
Remarks. Except for first periaxial cell cut off at apices of C. gasparrinii, each periaxial cells produce two cortical initials acropetally and one basipetally for a total of three initials (Fig. 4B). The first periaxial cell produces three cortical initials acropetally and one basipetally for a total of four initials. Each of the cortical initials after the first produces a descending uniseriate file of cortical cells, and together these completely cover internodal regions. C. gasparrinii is distinguished from all other congeners in Bermuda by its ovoid terminal acropetal cells (Fig. 5B), which are elongated in all of the other species, the small size of its spines, and its small rectangular cells in descending cortical filaments. Won et al. (2009) report whorled 3-celled spines at the nodes in C. gasparrinii but illustrate 2-celled spines (Won et al. 2009, Fig. 14b) similar to those found in the Bermuda specimens. Later, Won (2010) explained that he included the cortical initial as one of the cells of the spines. All of the other characteristics that Won et al. (2009) report for C. gasparrinii are in conformity with those found in Bermuda specimens.
It is not surprising that C. gasparrinii occurs in Bermuda, as it is presently the most geographically widespread of all of the 15 currently accepted species in the genus (Guiry and Guiry 2014). Using rbcL sequences, Won et al. (2009) identified this species from a number of western Atlantic locations from Brazil to Mexico and Florida, but report a range that also includes the Mediterranean Sea, the Indian Ocean, Asia, South Africa, Hawaii, Pacific Central America, and California. It seems likely that C. gasparrinii represents many of the specimens identified in the past as C. clavulatum giving it its former worldwide distribution. Bermuda specimens in the Collins et al. (1913) exsiccata, Phycotheca Boreali-Americana, distributed as no. 1948, Ceramium clavulatum C. Agardh, are C. gasparrinii. An earlier P.B.-A. collection distributed as C. clavulatum from Key West, Florida (Collins et al. 1895, No. 148a) likewise represents C. gasparrinii.
Description. Plants beige-pink with prostrate and erect axes 110-170 µm in diam. and to 6 cm tall; branching pattern pseudodichotomous often with a branch occurring in the notches of main branches at apices, branches forming at intervals of 8-15 axial cells on the main axes, the branches erect with forcipate apices (Fig. 6A); adventitious branches developing from periaxial cells in lower portions; uniseriate, multicellular rhizoids issued from prostrate axes and lower portions of erect axes; 6-10 straight, 2-celled spines whorled at axial cell nodes, one almost always found in notches of dichotomous branches, 15-18 µm diam. and 40-68 µm long (Fig. 6B); distal ends of axial cells cutting off 10-13 periaxial cells, these remaining at the nodes during axial cell elongation (Fig. 6C); periaxial cells cutting off two cortical initials acropetally and one basipetally (Fig. 4C); the first cortical initial cutting off one spine and two elongate cortical cells, one gland cell and two elongate cortical cells, or three elongate cortical cells, the second cortical initial cutting off one elongate acropetal cortical cell and one basipetal cortical filament, the third cortical initial cutting off one basipetal filament; basipetal filaments made up of mostly staggered cortical cell files of 7-11 cells, the cells elongate rectangular in surface view, the smallest 2-5 µm diam. and 21-28 µm long, the largest 5-10 µm diam. and 19-33 µm long, these completely corticating axial cells from node to node (Fig. 6D); gland cells ovoid; tetrasporangia formed in whorls at the nodes, one produced from each periaxial cell, subspherical, 35-43 µm diam. and 38-55 µm long, subtended by 0-2 involucral filaments; gametangia not seen.
[Fig. 6.] Centroceras hyalacanthum. (A) Axis with branch forming in the notch of the main axis, CWS/CEL/TRP 13-16-5. (B) Axial node with cortical initials bearing elongated acropetal cells and spines, TRP 12-171-3. (C) Cross section of axis showing 14 periaxial cells, CWS/CEL 10-30-12. (D) Axial segment displaying staggered, basipetal filaments of cortical cells, TRP 12-171-3. Ax, axial cell; C1 & 2, sequence of cortical initials; CEL, Christopher E. Lane; CWS, Craig W. Schneider; P, periaxial cell; TRP, Thea R. Popolizio. Scale bars represent: A, 1 mm; B & D, 25 μm; C, 50 μm.
Type locality. Antilles (French West Indies), Caribbean Sea.
Distribution. Bermuda, Florida, Caribbean Sea.
Selected collections. Bermuda—CWS/CEL 09-5-9, CWS/CEL 10-27-2 [BDA0428], CWS/CEL 10-30-12b [BDA0462], CWS/CEL/TRP 12-10-13 [BDA0590], CWS/CEL/TRP 12-18-15 [BDA0653], TRP 12-171-3 [BDA1810]. Florida, USA—CWS/CEL/TRP 13-8-17 [KW090], CWS/CEL/TRP 13-16-5 [KW265] (for collection details see Table 1).
Remarks. Multiple Bermuda isolates clustered with Florida, USA and Caribbean sequences of C. hyalacanthum in our COI-5P and rbcL trees (Figs 1 & 2). In C. hyalacanthum, the first periaxial cell cuts off three cortical initials acropetally and one cortical initial basipetally for a total of four cortical initials. On the rest of the periaxial cells, only three cortical initials form, two acropetally and one basipetally (Fig. 4C). In all cortical initial groups, the first cortical initial cuts off one spine and two elongate cortical cells, one gland cell and two elongate cortical cells, or three elongate cortical cells. This production of cortical cells from acropetal cortical initials is unique among all of the species thus far isolated from Bermuda (Figs 4C & 6B), the rest producing only two cells from their first cortica&l initials. In addition, descending cortical cell files are made of elongate rectangular cells (Fig. 6D). This developmental pattern of the cortex initiated at the nodes is in perfect accord with that demonstrated by Won et al. (2009) for the species including their observations on the lectotype specimen they designated. Amongst all of the species in the genus found in Bermuda, C. hyalacanthum has the narrowest axes, 110-170 µm in diam. (Table 2), the most delicate, from Florida and the Caribbean, being even narrower than those reported by Won et al. (2009), at 140-165 µm in diam.
In the same paper in which Kützing (1842) wrote the protologue for C. hyalacanthum, he also described an additional Caribbean species, C. oxyacanthum Kütz. (type locality = Cuba), and later described C. brachyacanthum Kütz. (type locality = Antilles, West Indies) (Kützing 1863). After an examination of types of all three species, Won et al. (2009) considered the latter two species to be heterotypic synonyms of C. hyalacanthum.
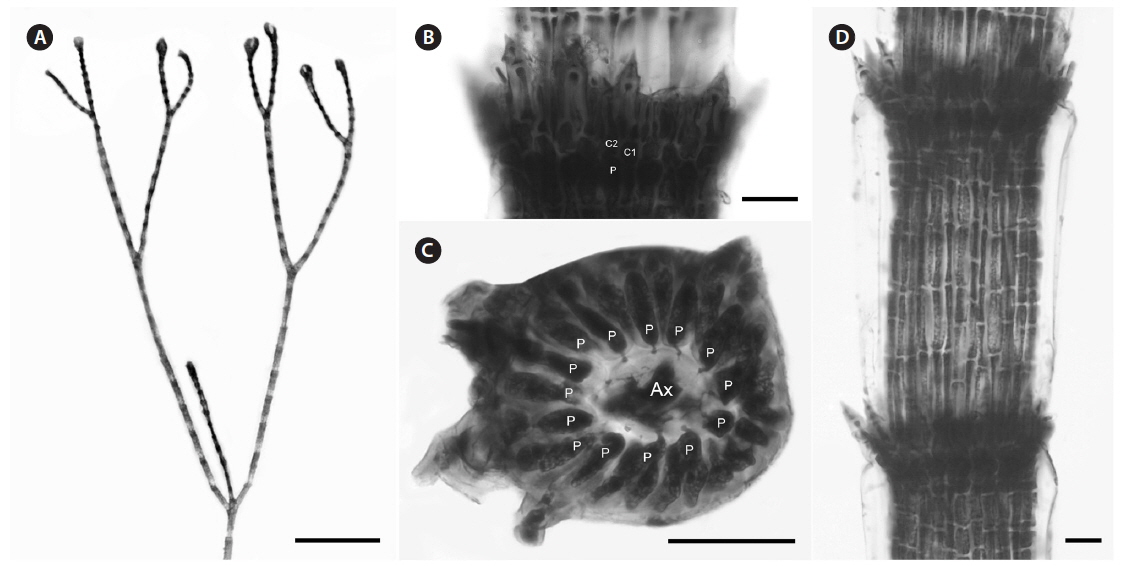
Description. Plants light rosy-red, with prostrate and erect axes 140-210 µm in diam. and to 2 cm tall (Fig. 7A); branching pattern pseudodichotomous, rarely with a branch forming in the notches of main branches at apices (Fig. 7B); branches forming at intervals of 11-12 axial cells on the main axes, branches erect with straight to slightly forcipate apices (Fig. 7B); adventitious branches occasionally developing from periaxial cells in lower portions; uniseriate, multicellular rhizoids issued in profusion from prostrate axes and lower portions of erect axes; 0-3 straight, 2-celled spines formed at axial cell nodes and in the notches of branches, one almost always found in notches of dichotomous branches, 12-14 µm diam. and 33-39 µm long (Fig. 7D); distal ends of axial cells cutting off 10-12 periaxial cells, these remaining at the nodes during axial cell elongation (Fig. 7C); except for the first abaxial periaxial cell, periaxial cells cutting off two cortical initials acropetally and one basipetally (Fig. 4D); the first cortical initial cutting off one spine and one elongate cortical cell, one gland cell and one elongate cortical cell, or two elongate cortical cells; the second cortical initial cutting off one elongate acropetal cortical cell and one basipetal cortical filament; the third cortical initial cutting off one basipetal filament; basipetal filaments made up of 8-9 cortical cells in tiered files, the cells isodiametric to short rectangular in surface view, the smallest 7-8 µm diam. and 10-13 µm long, the largest 5-14 µm diam. and 20-25 µm long, these completely corticating axial cells from node to node (Fig. 7D & E); gland cells ovoid; gametangia and tetrasporangia not seen.
[Fig. 7.] Centroceras illaqueans sp. nov. (A) Habit, holotype, CWS/CEL 09-2-9. (B) Axis with straight, pseudodichotomous branching, CWS/CEL 09- 2-9. (C) Cross section of axis showing 10 periaxial cells, CWS/CEL 09-2-9. (D) Axial node with cortical initials bearing elongated acropetal cells and single spine (arrowhead), CWS/CEL 09-2-9. (E) Axial segments displaying tiered, basipetal filaments of isodiametric cortical cells and lack of spines at nodes, CWS/CEL 09-2-9. Ax, axial cell; C1 & 2, sequence of cortical initials; CEL, Christopher E. Lane; CWS, Craig W. Schneider; P, periaxial cell. Scale bars represent: A, 2 cm; B, 1 mm; C-E, 25 μm.
Diagnosis. Differing from other species in the genus by its unique COI-5P barcode, and from the morphologically similar Centroceras hommersandii Won, T. O. Cho et Fredericq (2010) by producing more than one elongated acropetal cell from the cortical cell units.
Etymology. illaqueans (L., part., n.) = entrapping, for the habit of the new species sifting and affixing sand among its erect filaments in intertidal pools.
Holotype (designated here). CWS/CEL 09-2-9, Mar 15, 2009, tidal pool, rocky point south of beach, Capt. Williams’ Bay, Bermuda Is., 32°18′09.3″ N, 64°44′22.4″ W, Bermuda, western Atlantic Ocean [MICH] (Fig. 7A), GenBank Nos. KP222781, KP222802; isotypes—KIRI, NY, US, Herb. CWS.
Distribution. Endemic to Bermuda and currently known only from the type locality.
Remarks. C. illaqueans is genetically distinct from all the other sequences in our analyses, and is grouped with C. hyalacanthum in our trees (Figs 1 & 2). The new species is distinguished from other Centroceras species in Bermuda by the length of its terminal elongated acropetal cells which are mostly equal to or longer than spines at the nodes (Table 2, Fig. 7D & E), and the alignment of its basipetal filaments which typically line up with other descending files in tiers of short isodiametric cells (Fig. 7D & E). In addition, the new species differs from C. hyalacanthum by its fewer number of spines at the nodes, spine length and acropetal cell length (Table 2). A recently described species from Natal, South Africa, C. hommersandii, shares its small size with C. illaqueans, but has axes that are narrower in diameter than the Bermuda species and produces only a single elongated acropetal cortical cell in each cortical cell unit, being produced from the second cortical initial (Won et al. 2010). The first cortical initial of C. hommersandii produces a spine or gland cell and an ovoid acropetal cortical cell (Won et al. 2010), unlike the elongated cell produced by C. illaqueans.
Despite the numerous collections of Centroceras sequenced from Bermuda, C. illaqueans was found only once. In this extensive population, the tips of erect axes emerged approximately one half centimeter out of sandy sediment on the bottom of a large, shallow tidal pool on the south shore. Repeated visits to the type locality have not uncovered this species again.
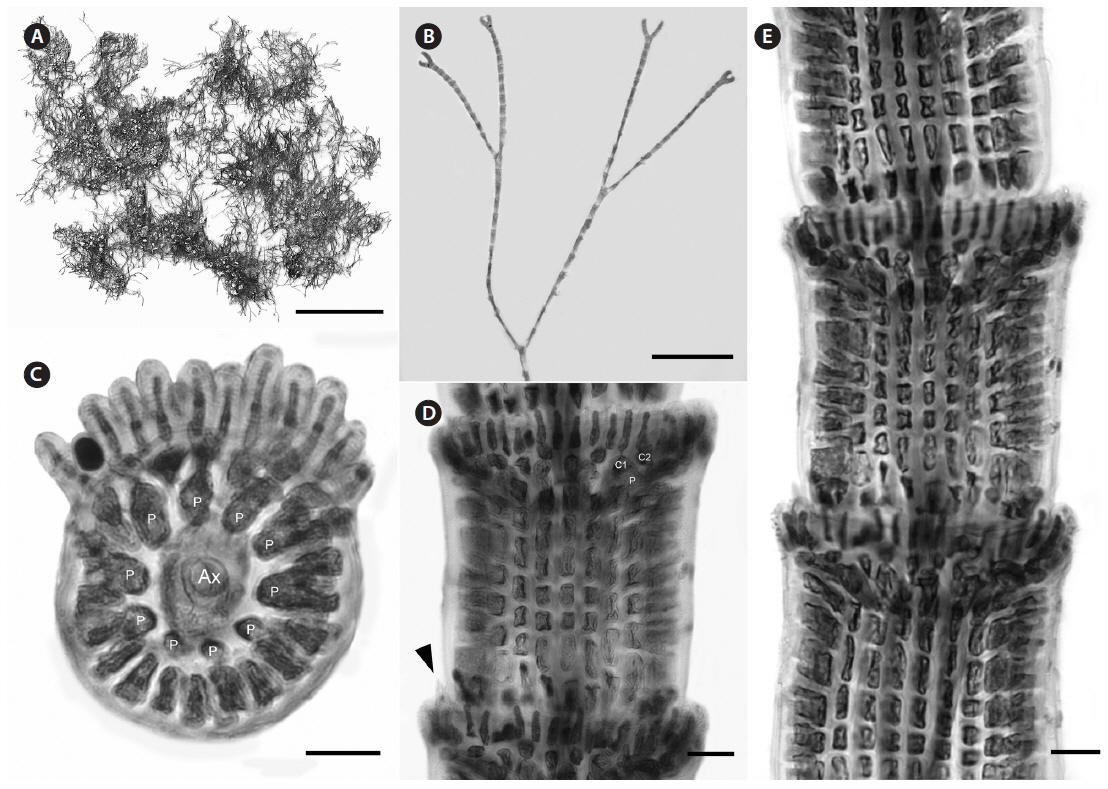
Description. Plants pink to dark rosy-red, with prostrate and erect axes 130-240 µm in diam. and to 2-4 cm tall; branching pattern pseudodichotomous, branches forming at intervals of 9-12 axial cells on the main axes, the branches divaricate with forcipate apices, some arched reflexive (Fig. 8A); adventitious branches developing from periaxial cells in the lower portions; uniseriate, multicellular rhizoids issued from prostrate axes and lower portions of erect axes; 2-8 straight, 2-celled spines whorled at axial cell nodes, one almost always found in notches of dichotomous branches, 15-40 µm diam. and 48-118 µm long (Fig. 8B-D); distal ends of axial cells cutting off 14-16 periaxial cells, these remaining at the nodes during axial cell elongation (Fig. 8C); periaxial cells cutting off two cortical initials acropetally and one basipetally (Fig. 4E); the first cortical initial cutting off one spine and one elongate cortical cell, one gland cell and one elongate cortical cell, or two elongate cortical cells, the second cortical initial cutting off one elongate acropetal cortical cell and one basipetal cortical filament, the third cortical initial cutting off one basipetal filament; basipetal filaments made up of mostly staggered cortical cell files of 5-17 cells, the cells elongate rectangular in surface view, highly variable in size, from 2-12 µm diam. and 14-116 µm long, these completely corticating axial cells from node to node (Fig. 8D), at times these cells lined up in tiers; gland cells ovoid; gametangia and tetrasporangia not seen in Bermuda collections. This species is best distinguished from C. arcii by its COI-5P barcode sequence (see Table 1 for GenBank numbers).
[Fig. 8.] Centroceras micracanthum. (A) Axis with reflexive, pseudodichotomous branching, CWS/CEL/TRP 13-16-25. (B) Axial node with cortical initials bearing elongated acropetal cells and spines, CWS/CEL/TRP 12-10-9. (C) Cross section of axis showing 15 periaxial cells, CWS/CEL/TRP 12-11-12. (D) Axial segment displaying staggered, basipetal filaments of cortical cells, CWS/CEL/TRP 12-10-9. Ax, axial cell; C1 & 2, sequence of cortical initials; CEL, Christopher E. Lane; CWS, Craig W. Schneider; P, periaxial cell; TRP, Thea R. Popolizio. Scale bars represent: A, 2 mm; B, 25 μm; C & D, 50 μm.
Type locality. Rio de Janeiro, Brazil, western Atlantic Ocean.
Distribution. Brazil, Caribbean, Bermuda, Cape Verde, Mediterranean, Polynesia.
Selected collections. Bermuda—CWS/CEL 09-16- 5, CWS/CEL 10-11-4a [BDA0084], CWS/CEL 10-14-24 [BDA0179], CWS/CEL 10-30-12a [BDA0460], CWS 12-10-9 [BDA0583], CWS/CEL/TRP 12-11-12 [BDA0611], CWS/CEL/TRP 12-18-14 [BDA0652], CWS/CEL/TRP 12-19-2 [BDA0658], TRP 12-28-2 [BDA0778], TRP 12-46-3 [BDA0971], TRP 12-59-3 [BDA1075], TRP 12-134-4 [BDA1561], TRP 12-153-6 [BDA1682], TRP 12-169-1 [BDA1795], TRP 12-173-9 [BDA1825]. Florida, USA—CWS/CEL/TRP 13-6-29 [KW039], CWS/CEL/TRP 13-16-25 [KW288] (for collection details see Table 1).
Remarks. In our rbcL analysis, sequences isolated from Bermuda specimens grouped with C. micracanthum from Florida and the Caribbean (Fig. 2), and our morphological characters matched those described from the same areas (Won et al. 2009). In the same work with the protologue of C. micracanthum, Kützing (1842) also described C. leptacanthum Kütz. (type locality = Genoa, Italy), C. cryptacanthum Kütz. (type locality = Antilles, West Indies) and C. macracanthum Kütz. (type locality = Brazil), all of which are now considered heterotypic synonyms of C. micracanthum after an examination of type material (Won et al. 2009). Given its widespread distribution in the western Atlantic (Guiry and Guiry 2014), it is not surprising that C. micracanthum is found in Bermuda with many other macroalgal species also known in the Caribbean. Based upon morphological characteristics, Florida specimens of C. clavulatum distributed as No. 1347 in the Collins et al. (1906) exsiccatae, Phycotheca Boreali-Americana, are better identified as C. micracanthum. The label noted that these Key West specimens correspond to C. leptacanthum, a species that Won et al. (2009) maintained under C. micracanthum as noted above.
As discussed above, C. micracanthum is most difficult to distinguish morphologically from C. arcii in Bermuda. Its habit of reflexive branching in about 75% of specimens and overlap of anatomical characteristics make it truly cryptic with C. arcii. Therefore, this species is best identified by comparing genetic sequences with those available in the public domain.
The use of molecular-assisted alpha taxonomy has allowed for the resolution of five species in the ‘Centroceras clavulatum complex’ in Bermuda. Won et al. (2009, Fig. 14) used stylized diagrams of nodal cortical units for each of the species they covered in their seminal study of this complex. They depicted the origin, number and shape of cortical cells, glands and spines produced from cortical initials as distinguishing features among the species, and we have done the same for nodal cortical initials bearing spines (Fig. 4), and the developmental patterns of our two new species can be compared in this way. Three of the five Bermudian species are recorded in their most northerly locations in the western Atlantic, and the remaining two, C. arcii and C. illaqueans, are newly described from the islands. Studies in other parts of the world’s warm temperate to tropical seas will undoubtedly extend or shrink the distributional ranges of some Centroceras species, and thousands of herbarium specimens in the genus world-wide will need further study and annotation. Only then will we have a more accurate picture of the biogeography of the many species in the genus.