Seawater contains diverse organisms and viruses that infect their host, with ~105-109 virus particles in 1 mL of seawater (Suttle et al. 1990, Suttle 2007). The majority of these viruses infect bacteria, cyanobacteria, archaea, and other eukaryotic organisms, but some of these viruses infect macro- or microalgae. These algal viruses are known to play an ecologically important role in regulating the population dynamics of their phytoplankton hosts (Suttle et al. 1990, Bratbak et al. 1993). Over 50 different viruses or virus like particles (VLPs) infecting marine eukaryotic algae have been isolated and characterized during the last two decades (Van Etten et al. 1991, Nagasaki and Bratbak 2009, Wilson et al. 2009).
Among the primary producers in aquatic ecosystems, diatoms comprise a major group of algae and are the most common phytoplankton. More than 105 species of diatoms are estimated to exist, which account for up to 35-75% of marine primary production in the ocean (Smetacek 1999). Therefore, lysis of these species by virus infection can reduce primary production. For example, the inoculation of phytoplankton communities of several major algal species with viral concentrates from seawater reduced primary production by as much as 78% (Suttle et al. 1990, Suttle 2007).
Despite the potent roles of diatoms and viruses infecting these hosts in aquatic ecosystems, few studies have examined diatom viruses. Since the first diatom virus was, at least nine diatom viruses have been isolated (Tomaru et al. 2011
In the present study, we isolated and characterized a novel virus infecting a diatom,
The host organism,
>
Isolation of virus particles
Seawater samples were collected at Jaran bay, Korea, between May and October 2008 and prefiltered through 0.8 µm pore-size polycarbone filters (Nuclepore, Pleasanton, CA, USA). The logarithmic phase cultures of
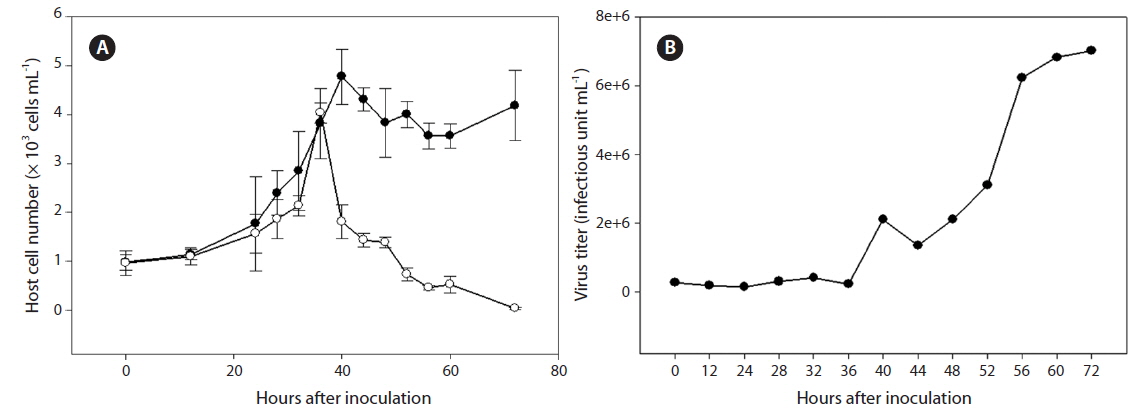
Cultures containing 500 mL of exponentially growing host cells were inoculated with 20 mL of lysate containing SpalV at a viral titer of 3.0 × 106 estimated by the most probable number; cell density and virus titer were then respectively measured using light microscopy and the extinction dilution method (Tarutani et al. 2001) every 24 h until 120 h postinoculation (hpi) (Fig. 2). And the host range of SpalV was examined by adding 50 µL of the original pathogen suspension to each 1 mL culture of exponentially growing algal strains listed in Table 1. Each culture was incubated under the culture conditions described above and observed under a light microscope.
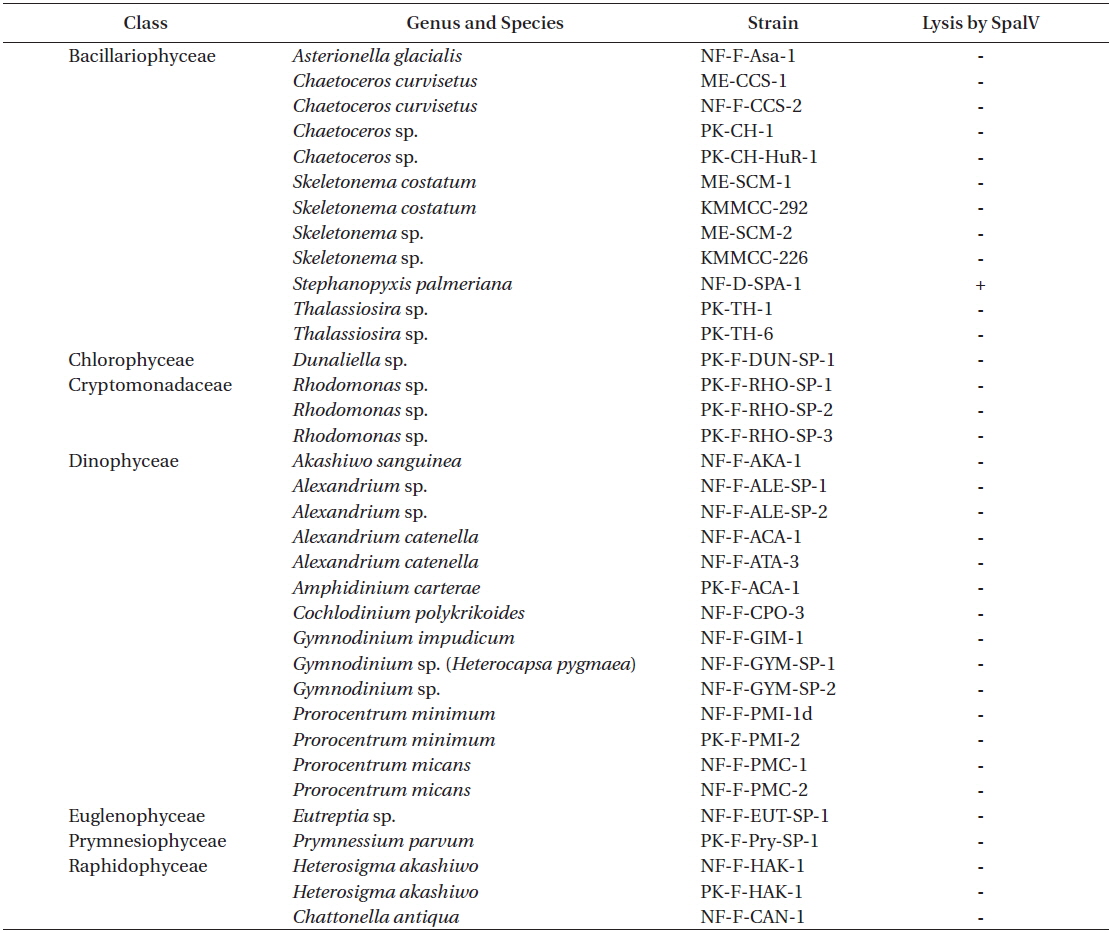
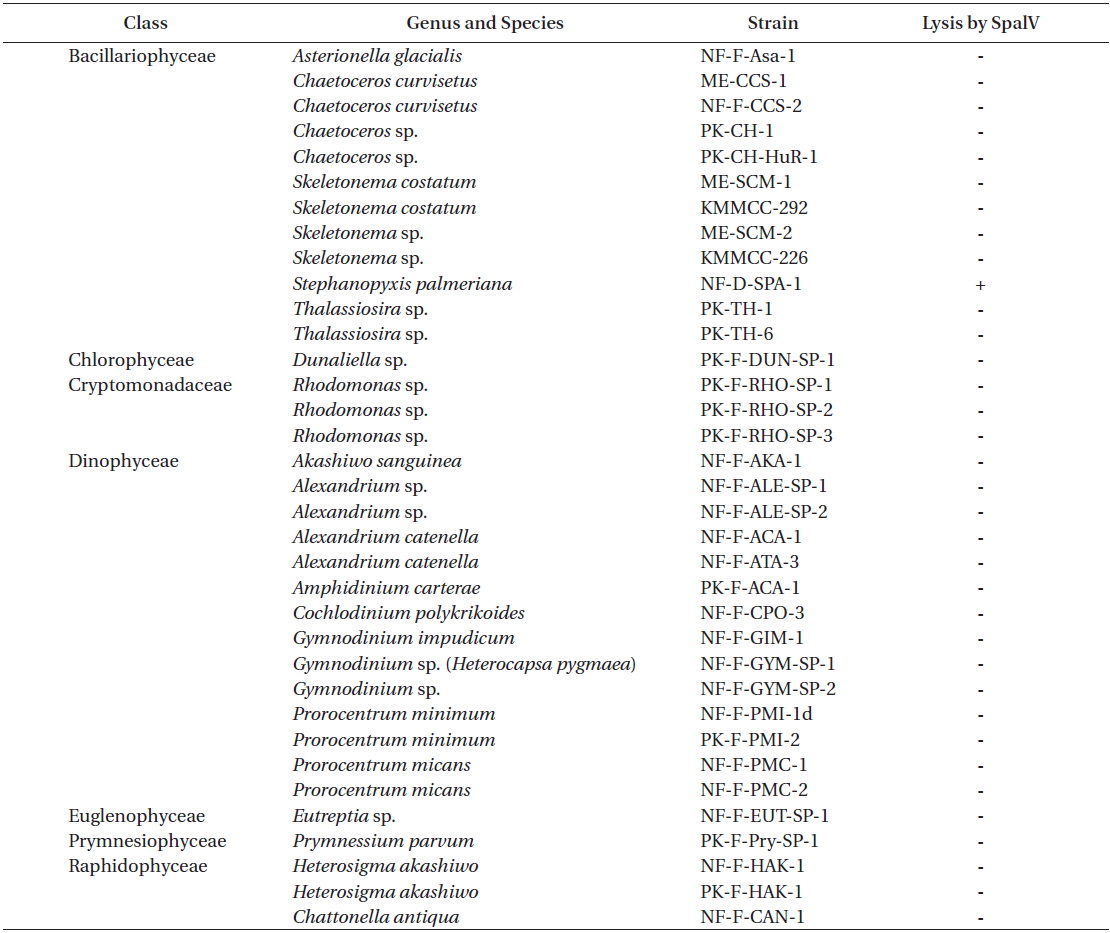
Infection specificity of Stephanopyxis palmeriana virus (SpalV) against 35 strains of marine phytoplankton
>
Transmission electron microscopy (TEM)
The morphology of SpalV was observed using a TEM.
The VLPs were also observed in negative staining with 4% uranyl acetate using a transmission electron microscope. Briefly, a drop of purified virus suspension was mounted on a grid for 30 s, and excess water was removed using filter paper. Then, a drop of 4% uranyl acetate was mounted on the grid for 10 s, and removed using filter paper. After the grid was dried, the grids were observed using TEM with the same conditions as above. Particle diameters were estimated using the negatively stained images.
The effect of storage temperature on SpalV infectivity was examined according to the method of Tomaru et al. (2005). An exponentially growing culture of
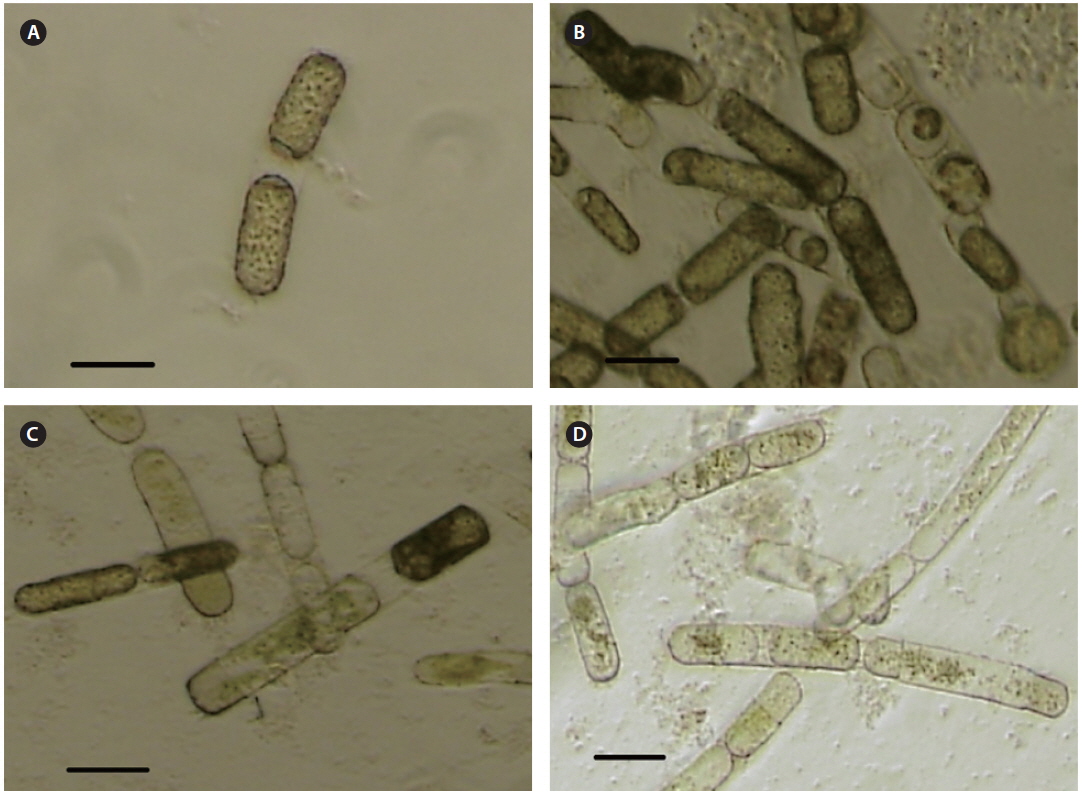
Cultures and cells of
SpalV was not lytic to any microalgal species tested other than
The replication parameters of SpalV were determined based on growth experiments. An increase in viral titer was detected from 36 to 56 hpi, and the lytic cycle of SpalV was predicted to be shorter than 40 h. Furthermore, by comparing the decrease in host cell number to the increase in viral titer between 48 and 80 hpi (Fig. 2A), the burst size was estimated to be 92 infectious units cell-1 (Fig. 2B). An increase in viral titer during exponential host growth has been commonly observed in
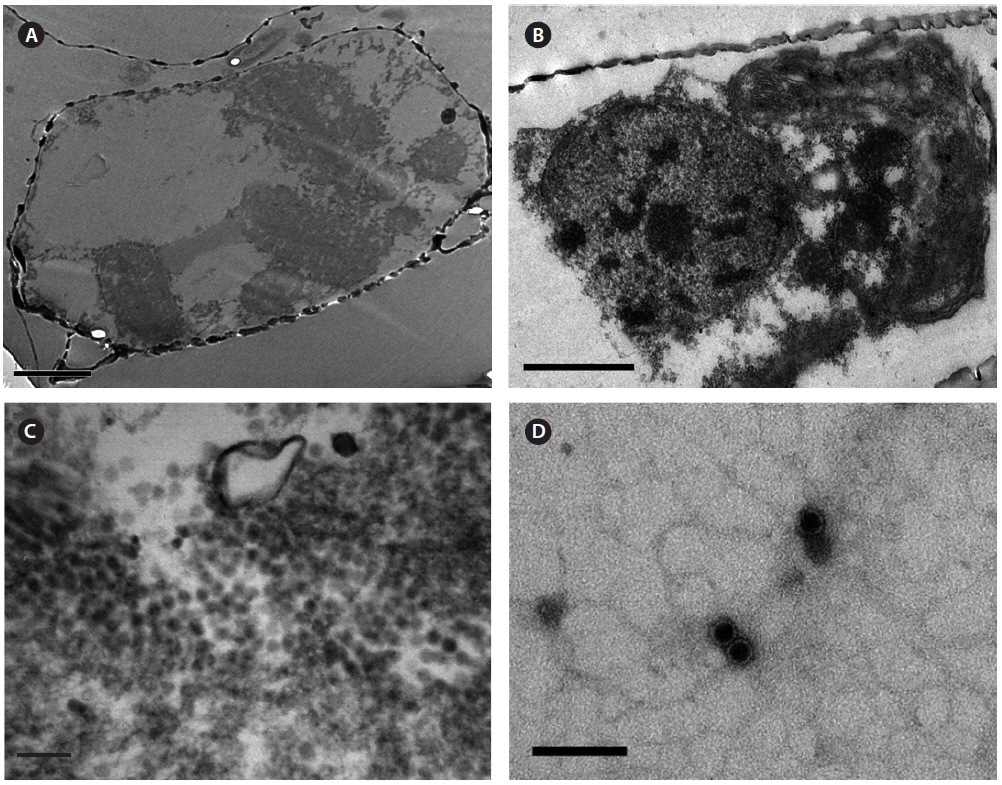
Intracellular virus particles were observed mainly in the cytoplasm, and the roundish virus-like particles were ~25-28 nm in diameter which were not observed in noninfected healthy host cell (Fig. 3A-C). No typical crystalline array formation reported in other diatom viruses was observed (Nagasaki et al. 2005, Tomaru et al. 2008, 2013). Virus particles observed by negative staining were 20 ± 5 nm in diameter, and no outer membrane or tail-like structures were observed (Fig. 3D). Similar structures in the virus-infected host cells have been reported for other diatom viruses, including viruses infecting
After 1 month of storage at 4, 15, and 20°C in the dark, the estimated titers were 7.02 × 104 , 1.9 × 104 , and 5.1 × 103 infectious units mL-1, respectively, but were below the detection limit (<3.0 × 10 infectious units mL-1) after 3 months of storage at each temperature. The decrease in infectious titers has been also reported for other microalgal viruses such as
Based on previous reports, we know that phytoplankton viruses smaller than 40 nm in diameter typically contain ssRNA genomes and are related to viruses from the highly diverse picorna-like superfamily (Culley et al. 2003). These are the cases for the ssRNA virus previously found in a diatom (
Viruses are thought to influence community composition through increased infection of competitively superior host species that are abundant and are fast growing (Sandaa 2008). Although we do not have sufficient data to determine the roles of viruses in marine environments or in host population dynamics, the interaction between
We have reported basic characteristics of a novel virus (
To the best of our knowledge, this is the first report of a diatom virus infecting and lysing