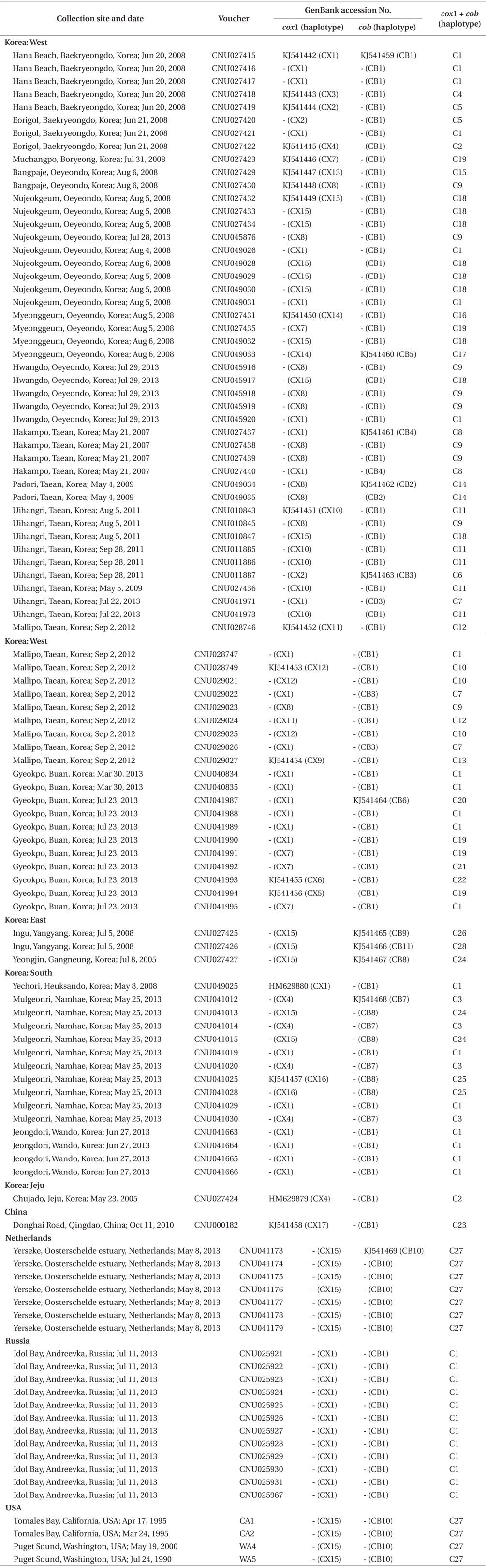
The number of species of non-native and invasive marine algae is growing, with concomitant public concern about native ecosystems and coastlines. One of the interesting biological questions is how these species are extending their natural ranges despite low genetic diversity (e.g., Jousson et al. 2000, Provan et al. 2005, Voisin et al. 2005, Kim et al. 2010). However, the genetic structure of native populations has been less intensively studied than that of the non-native ones, and may provide clues to the mechanism of invasion.
The red alga Gelidium vagum Okamura commonly occurs in Korea, Japan, China and far eastern Russia (Kang 1966, Zhang and Xia 1988, Yoshida 1998, Kozhenkova 2009). The species was first described on the basis of intertidal specimens from western coast of Hokkaido to Iwaki Province (Okamura 1934). The type locality, although not noted in the protologue (Okamura 1934), was later designated as Onahama Harbor in Fukushima Prefecture, Japan (Yoshida 1998). In Korea, G. vagum was first reported at Kiseong on the east coast, Odongdo in south, and Huksando in west (Kang 1966); its distribution has since been extended to Jeju and Ocheongdo (Sohn and Kang 1978, Lee 1988, 1994,Lee and Kim 1995).
Renfrew et al. (1989), relying morphological characters, reported the introduction of Gelidium vagum to British Columbia, Canada. Further introductions of this species in California and Washington, USA, and the Netherlands have been confirmed by comparing rbcL sequences (Hughey et al. 1996, Mineur et al. 2012). The Asian origin of G. vagum has been demonstrated by phylogenetic analyses of rbcL, and cox1 sequences (Boo et al. 2014).
Gelidium vagum serves as an excellent model system for the study of heterosis in red algae because fronds can readily be grown under laboratory conditions and the sexual reproductive cycle can be completed in a few weeks under optimum conditions (van der Meer and Patwary 1991, Patwary et al. 1993, Patwary and van der Meer 1994). Although its mitochondrial genome was fully analyzed (Yang et al. 2014), little information on the genetic diversity of G. vagum in its native range is available.
Mitochondrial-encoded cox1 is a fast-evolving gene that has recently proved valuable for barcoding red algal species (Saunders and Moore 2013) and for revealing the biogeographic structure of populations (Kim et al. 2010, 2012). Cytochrome b (cob), one of the 12 markers undergoing tests to determine its utility for phylogenetic studies by the Research Group for the Red Algal Tree of life (Saunders and Moore 2013), has potential for assessing genetic variation within species. The aim of this study was to contribute to an understanding of the phylogeography of this invasive alga by investigating haplotype and nucleotide diversities of both cox1 and cob genes in Gelidium vagum from Korea.
Specimens in this study are deposited in the Herbarium of Chungnam National University, Daejeon, Korea. A total of 108 individuals was collected in Korea, China, far eastern Russia, the Netherlands, and USA (Table 1). Field-collected specimens were desiccated in silica gel. Epiphytes on specimens were removed under a dissecting microscope before DNA extraction.
DNA extraction, polymerase chain reaction amplification, and sequencing procedures followed Boo et al. (2013). Specific primers for the amplification and sequencing were COXI43F and COXI1549R for cox1 (Geraldino et al. 2006) and CB44F (ATT TAA TWG ATT ATC CWA CDC C) and CB1006R (CAG GCA TWC CRC CWA TYC AKC) for cob (Saunders and Moore 2013). All sequences of the forward and reverse strands were determined for all taxa, and the electropherograms were edited using the program Chromas v.1.45 (Conor McCarthy, Griffith University, Southport, Queensland, Australia; http://www.technelysium.com.au/chromas.html) and rechecked manually for consistency. Nucleotide sequences were aligned using Se-Al v.2.0a11 (Rambaut 2002).
The minimum spanning network to analyze relationships and geographical distributions of each haplotype was constructed using Arlequin v.3.5.1.2 (Excoffier and Lischer 2010). Haplotype diversities (Hd) and nucleotide diversities (π) were measured using DnaSP v.5.10.01 (Librado and Rozas 2009).
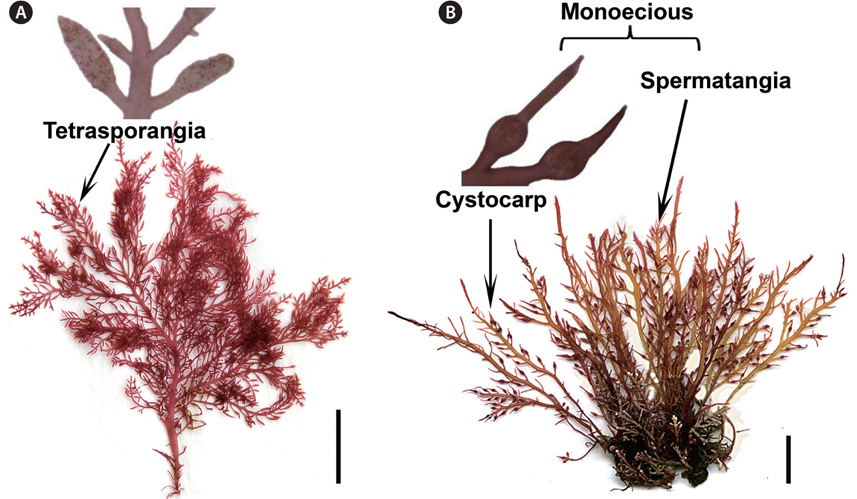
Our collections from Korea matched the morphological features of Gelidium vagum from Russia (Fig. 1A & B), the Netherlands and USA. Thalli are erect, up to 10 cm in height, pinnately to irregularly branched, with broad but very thin main axes and branches. The cortex is composed of round cells, and the medulla consists of a network of elongate cells with a few rhizoidal filaments in the outer medulla (figure not shown). Tetrasporangial stichidia, formed on terminal or lateral branches (Fig. 1A), are compressed and stipitate, with rounded tips. Tetrasporangia are in regular, acropetally developed rows, and are decussately or cruciately divided. Gametophytes are monoecious, forming cystocarps and spermatangia on the same individuals (Fig. 1B).
Thalli grew on rock or on the gastropod, Acmaea pallida at Oeyeondo Island (Fig. 2), in intertidal and subtidal zones, and sometimes were abundant on shaded rocks. Tetrasporangial and gametangial thalli were found from June to August in Korea and eastern Russia.
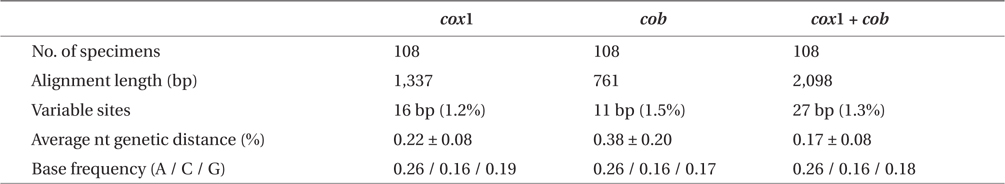
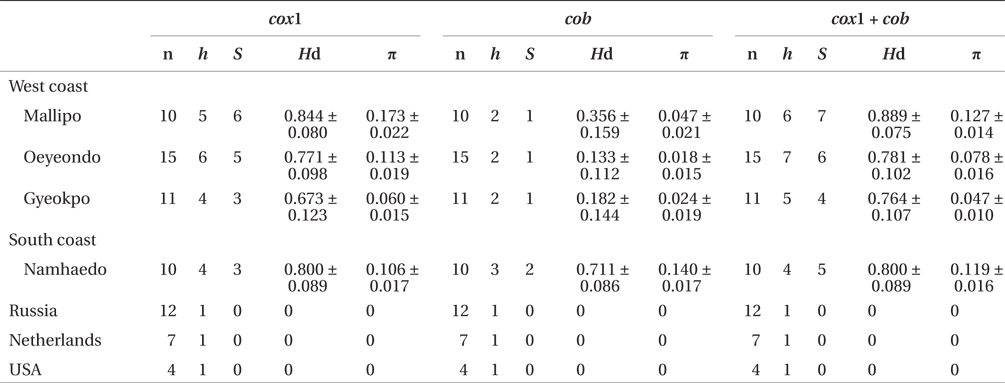
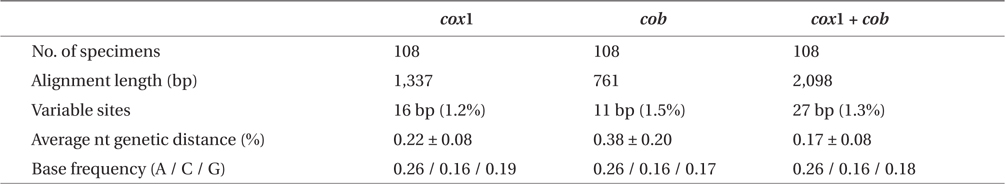
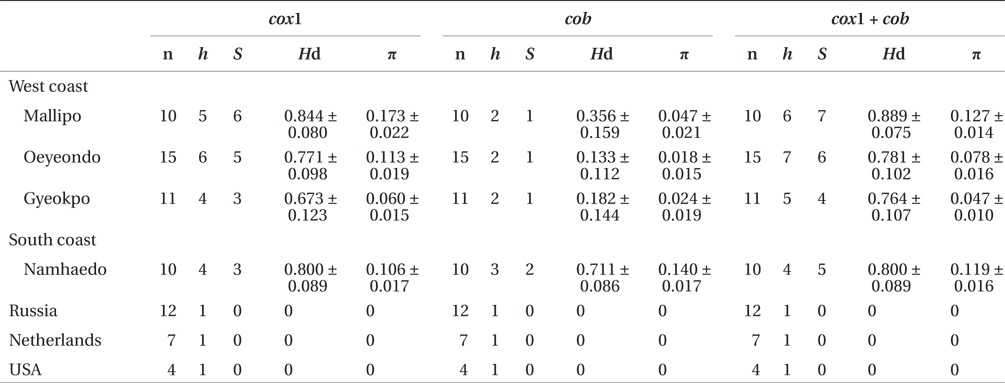
Sequence comparisons of the 1,337 base pair (bp) cox1 fragment from 108 individuals revealed 16 polymorphic sites (1.2%), defining 17 haplotypes (Table 2). Ten haplotypes (59%) were ‘private’ (unique to a single region). Haplotype and nucleotide diversities of cox1 in the Mallipo population (n = 10) on the west coast were high (Hd, 0.844; π, 0.173), while both indices were relatively low in Gyeokpo (Hd, 0.673; π, 0.060). In the Russian population (n = 12), both diversities were zero (Table 3). Haplotype network of cox1 revealed no genetic structure in Korean populations of G. vagum, suggesting genetic flow among local populations of the species (Appendix 1). Haplotype 15 occurred in Yerseke, the Netherlands, Tomales Bay and Puget Sound, USA, as well as several places in Korea (Table 1).
Sequence comparisons of the 761 bp cob fragment from 108 individuals revealed 11 polymorphic sites (1.5%), defining 11 haplotypes (Table 2). Haplotype and nucleotide diversities of cox1 coast were high in Namhaedo (Hd, 0.711; π, 0.140), whereas both indices were low in Oeyeondo (Hd, 0.133; π, 0.018) and in Gyeokpo (Hd, 0.182; π, 0.024). Russian population (n = 12) was zero for both diversities (Table 3). Although the number of cob haplotypes was smaller than that of cox1 haplotypes, the network of cob haplotypes (Appendix 2) is similar to that of cox1, showing no genetic structure in Korean populations. Haplotype CB10 occurred in the Netherlands and USA (Table 1).
The combined cox1 + cob dataset (2,098 bp) revealed 27 polymorphic sites (1.3%), defining 28 haplotypes. The number of haplotypes was high on the west coast (32%, 21 haplotypes / 65 individuals) and relatively low on the south (26%, 4 / 15). Haplotype and nucleotide diversities were high in Mallipo (Hd, 0.889; π, 0.127), while both indices were relatively low in Oeyeondo (Hd, 0.781; π, 0.078) and Gyeokpo (Hd, 0.764; π, 0.047). Both diversities in the Russian population (n = 12) were zero (Table 3).
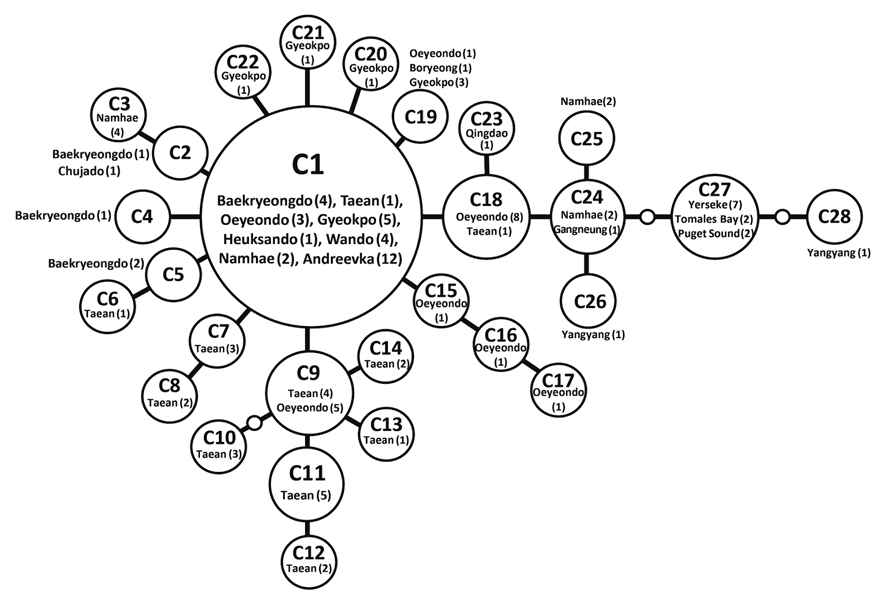
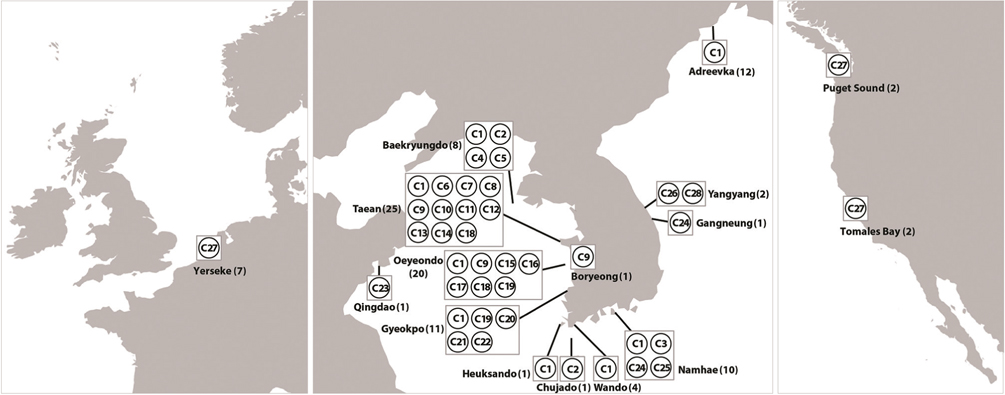
Haplotype network of cox1 + cob (Fig. 3) was similar to that of individual networks, showing no genetic structure in Korean populations of G. vagum. Haplotype C1, placed in the center of a star-like structure, commonly occurred on the west and south coasts of Korea (Fig. 4). All specimens from Russia belonged to haplotype C1. Haplotype C27 occurred in the Netherlands and USA, but not in northeast Asian waters.
Gelidium vagum, a monoecious species (Renfrew et al. 1989, van der Meer and Patwary 1991), may be capable of selfing, resulting in reduced genetic variation within populations. However, Korean populations revealed a high number of haplotypes in cox1 (16%, 17 / 108 individuals), cob (10%, 11 / 108 individuals), and the combined cox1 + cob (26%, 28 / 108 individuals). Similarly, genetic diversities of cox1 (Hd, 0.809 ± 0.026; π, 0.109 ± 0.009) for G. vagum were relatively high. Our results are consistent with previous reports (Patwary et al. 1993) that G. vagum isolates have sufficient genetic diversity for conducting a heterosis experiment using random amplified polymorphic DNA methods. In contrast, genetic diversities were relatively low (Hd, 0.711) in G. elegans (Kim et al. 2012) and (Hd, 0.757) in Gracilaria vermiculophylla (Ohmi) Papenfuss (Kim et al. 2010). The latter two species are dioecious, species, a trait which may account for increasing heterozygosity and variability. However, because red algae may often reproduce asexually, the relationship between genetic diversity and monoecy or dioecy is not clear within natural populations.
Recent studies on the genetic diversity of introduced marine algae have revealed invasive haplotype(s) that are shared between native and introduced populations (Jousson et al. 2000, Provan et al. 2005, Voisin et al. 2005, Uwai et al. 2006, Kim et al. 2010). Our cox1 dataset suggests that the introductions of Gelidium vagum to Europe and north America originated from populations in Korea and / or Japan, as shown in previous morphological and rbcL studies (Renfrew et al. 1989, Hughey et al. 1996, Mineur et al. 2012). In addition, the cox1 dataset revealed that the haplotype CX15, shared by populations in Korea, USA, and the Netherlands, is an invasive haplotype. However, because marine algal introductions might have occurred more than once (Voisin et al. 2005, Hughey et al. 2009), further sampling to search for other invasive haplotypes is necessary to determine whether G. vagum was introduced via single or multiple events.
The number of private haplotypes and both haplotype and nucleotide diversities were higher on the west coast of Korea than on the other coasts. Haplotype C27 was found in the Netherlands and USA, where haplotype and genetic diversities were zero. Our results corroborate the generalization that the genetic diversity of invasive species is low in introduced populations relative to that in native populations. Because the invasive haplotypes identified here, for example, CB10 in cob and C27 in cob + cox1, were not found in Korea, we cannot identify a specific donor region in the combined data without further study.
The vector for introduction of Gelidium vagum to Canada, USA, and the Netherlands may be the import of oysters, as shown in many cases of invasive macroalgae elsewhere (e.g., Voisin et al. 2005, Miller et al. 2011), because the Hornby Island-Denman Island area, Canada and Oosterschelde estuary, the Netherlands, are hotspots for oyster aquaculture (Renfrew et al. 1989, Mineur et al. 2012). Other mollusks may prove to be vectors because G. vagum is often found on Acmaea pallida, as seen in Fig. 2. Acmaea pallida is commonly found on the hulls of ships, and may survive through long voyages.
This is the first report to analyze the mitochondrial cob gene as a molecular marker for red algae. The genetic distance values of cob (1.5%) were similar to those of cox1 (1.2%) (Table 2) and both demonstrated similar population structures for G. vagum (Appendices 1 & 2). In addition, our use of cob and cox1 genes reveals a very high number of haplotypes, compared to that of each individual gene, and thus increases the resolution of our haplotype network for G. vagum. The cob gene is a new marker for population studies of red algae, and its use can be extended to studies of other biogeographically interesting species of benthic algae. The analysis of more than one gene provides a more accurate picture of genetic diversity in marine algae and key information for conservation.