Bumblebees provides valuable pollination services to a wide variety of food crops and wild flowers, and thus the decline of bumblebee pollinators is of great concern for both ecological and economic reasons (Whittington and Winston, 2003). The following species of bumblebees have been used commercially for pollination around the world: Bombus terrestris, Bombus lucorum, Bombus occidentalis, Bombus ignitus, Bombus hypocrita sapporoensis and Bombus impatiens (Velthuis and van Doorn, 2006; Li et al., 2012). According to Weiser’s article (Weiser, 1961), the microsporidium described in bumblebees is identical to the parasite of the honey bee, which made Nosema bombi a junior synonym of Nosema apis (Larsson, 2007). Furthermore, common correlates of declining species include high N. bombi prevalence and intensity, and decreased genetic diversity (Koch and Strange, 2012). Microsporidia are obligate intracellular parasites of eukaryotes: outside another cell they can only exist as infective spores that are protected by walls of protein and chitin. Their distinguishing features are small size, thick walls, and the presence of an organelle called the polar tube. This long, thin tube lies tightly coiled within the cytoplasm of the spore, contacting the thinner point of the spore’s coat at its anterior end. Microsporidia damage apiaries, fisheries, and silk farms and cause severe disease in immuno-compromised humans (Keeling and Fast, 2002). N. bombi as an intracellular pathogen produces systemic infection in bumblebees (Koch and Strange 2012). N. bombi infects a number of different host tissues, which may explain why infection in multiple bumblebee species is observed (Fries, 2010; Shafer et al., 2009), and is suggested to have facilitated the collapse of commercial B. occidentalis populations in rearing facilities (Flanders et al., 2003). On the other hand, according to Schmid-Hempel’s article (Schmid- Hempel, 2001), parasite continuously adapt to the prevailing host types and hosts continuously escape this adaption by changing their genotypes, as a result, microsporidia- infected bumblebees generally produce more sexual offspring than uninfected colonies (Schmid-Hempel, 2001). The pathogen has “spilled over” into adjacent wild populations and is transmitted along with other pathogens (Bollan et al., 2013; Colla et al., 2006; Kissinger et al., 2011; Whittington and Winston, 2003) from commercial operations. N. bombi infects multiple bumblebee species (Shafer et al., 2009), and N. apis and N. ceranae are known from the Western (Apis mellifera) and Asiatic (Apis Cerana) honey bee, respectively (Fries et al., 1996). N. bombi may cause sluggish behavior and early death (Schmid-Hempel and Loosli 1998), whereas N. apis can be virulent to Western honey bees in temperate climates (Moeller, 1978). The morphological and genetic characteristics of N. apis are demonstrated to be closer to N. bombi (Vossbrinck and Debrunner-Vossbrinck, 2005), and N. bombi closer to N. ceranae (Shafer et al., 2009). Nosema disease causes extensive loss of adult bees in Korea and is responsible for some supersedure of queens (Choi et al., 2006). Therefore, for the protection of wild bees and to help prevent the further spread of Nosema disease, a rapid and effective diagnosis is obligatorily needed.
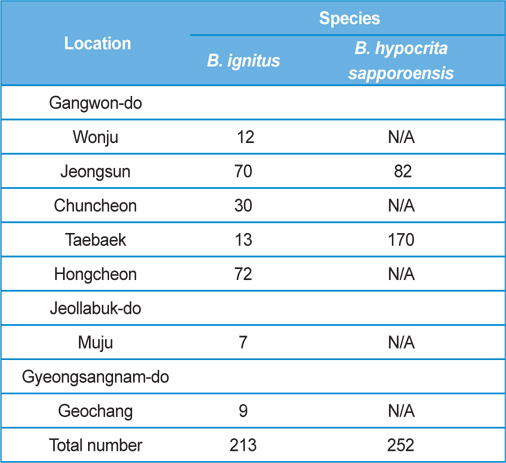
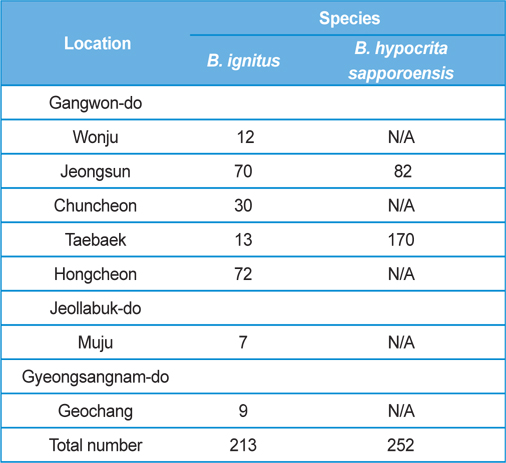
The post-hibernated queens of the Korean native bumblebees, B. ignitus and B. hypocrite sapporoensis, which mainly visit the blossoms of Corydalis speciosa (Fumariaceae) and Prunus × yedoensis (Rosaceae), were collected from numerous localities, including Wonju, Jeongsun, Chuncheon, Taebaek, and Hongcheon in Gangwon Province, Korea, during the spring season of 2013 (Table 1). The queens collected in the field were confined in round plastic containers (7.5 cm in diameter and 5.5 cm in depth; two or three queens per container) and supplied with 40 - 50% sugar solution until they moved into the nest initiation boxes. The duration from collection to the start of rearing was 1 d or less. The number of B. ignitus and B. hypocrita sapporoensis collected in the spring season of 2013 was 213 and 252, respectively.
The collected queens were reared in three types of cardboard boxes (1.5 mm thick), for nest initiation (10.5 × 14.5 × 6.5 cm), colony foundation (21.0 × 21.0 × 15.0 cm), and colony maturation (24.0 × 27.0 × 18.0 cm). Each box had a wire net window on its lid for ventilation. The sizes of these windows were 5.5 × 6.5 cm, 7.0 × 14.0 cm, and 10.0 × 20.0 cm, respectively. Queens collected in the field were first confined individually in small boxes for colony initiation and remained there until oviposition. When the adults emerged from the first brood, the nest was transferred to a medium box for colony foundation and left there until the number of workers reached 50. The nest was then moved to the large box for further colony development with a 40 - 50% sugar solution and fresh pollen collected from an apiary (v:v = 1:1).
Colony foundation meant that more than 50 workers emerged. During the colony foundation time, bumblebee workers died suddenly over a period of 1 to 2 d. To identify and assay for Nosema infection, living bumblebees (B. ignitus and B. hypocrita sapporoensis) were collected from Wonju, Jeongseon, Chuncheon, Taebaek, and Hongcheon in Gangwon Province, Korea (Table 1.) The bumblebee samples from Wonju, Chuncheon, and Hongcheon did not show Nosema symptoms and negative reactions were detected together with nonspecific bands from PCR. However, bumblebees from Jeongsun and Taebaek had symptoms of Nosema disease, and for the separation of microsporidia, bumblebee samples from Jeongsun and Taebaek were homogenized in distilled water and filtered through gauze. The PCR band results of bumblebee samples from Jeongsun and Taebaek exhibited positive results.
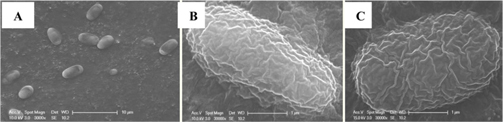
Newly identified microsporidian from bumblebees was named the JNK1. Adult bees infected with JNK1 and normal bees were dissected in distilled water in order to observe the microsporidian (Nosema species) structure by light microscopy. The sections for light microscopy were mounted onto slides and photographed. For the correct identification of the morphology of JNK1, we used electron microscopy. Sections for electron microscopy were mounted onto 200-mesh Ni grids, and viewed in a H-7000 Hitachi microscope at 10 kV and 7.5 kV. We observed the morphology of JNK1 by SEM after coating the grids with white gold.
DNA isolation, PCR amplification and sequencing of JNK1
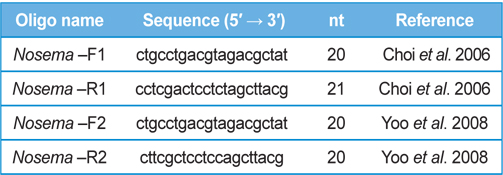
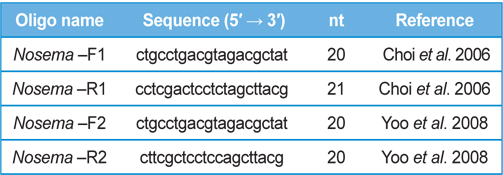
DNA from JNK1 was extracted from bumblebee samples suspended in an additional 500 μL of double-distilled H2O and centrifuged at 3000 rpm for 5 min. The supernatant was then discarded and the collected pellet was analyzed using a Wizard PCR Preps DNA Purification System (Promega, Madison, WI, USA). The 16S small subunit ribosomal RNA gene was amplified from each of the microsporidian isolates and amplified using the following two oligonucleotide primer pairs: Primer 1 Forward 5′- ctgcctgacgtagacgctat-3′ and Primer 1 Reverse 5′- cctcgactcctctagcttacg-3′ ; Primer 2 Forward 5′-ctgcctgacgtagacgctat-3′ and Primer 2 Reverse 5′- cttcgctcctccagcttacg-3′. These primers were used based on previous studies by (Choi et al., 2006; Yoo et al., 2008). PCR amplification was carried out in 20 μL, using 5 ng of DNA, 10 pmol of each primer, 0.2 mM of each deoxyribonucleotide triphosphate (dNTP), 2 mM of MgCl2, and 1 U of Taq Polymerase (Promega, USA). The amplification conditions for Primer pair 1 were as follows: 94℃ denaturation for 5 min, followed by 35 cycles of 94℃ for 30 s, 54℃ annealing for 30 sec, and extension at 72℃ for 1 min, with final extension of 10 min at 72℃ in a thermal cycler (Biometra, Germany). The conditions for Primer pair2 were as follows: 94℃ denaturation for 5 min, followed by 35 cycles of 94℃ for 20 s, 58℃ annealing for 20 s, and extension at 72℃ for 20 s, with final extension of 10 min at 72℃ in a thermal cycler. Sequence data were aligned using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) system and aligned using ClustalW in BioEdit.
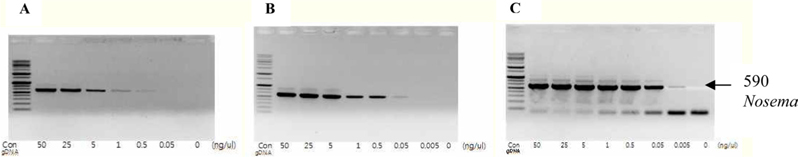
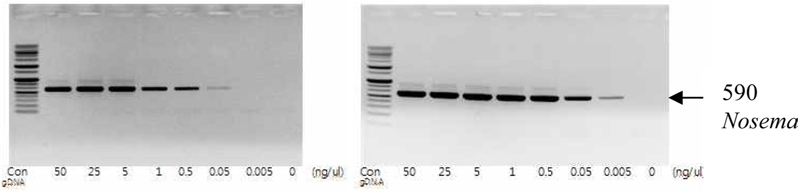
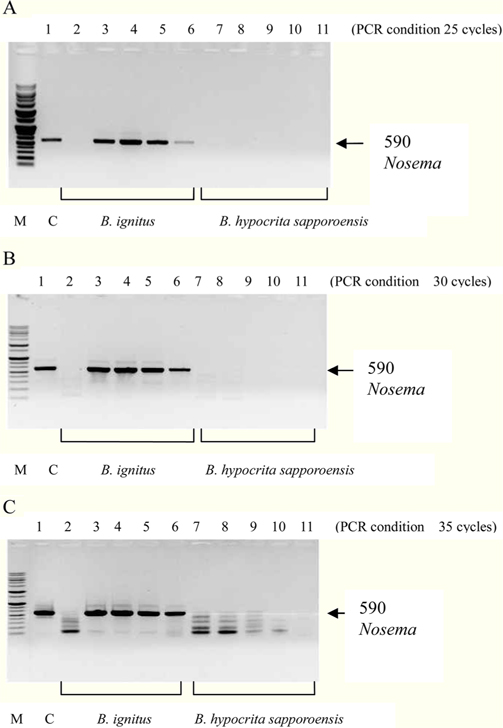
To detect the sensitivity of the PCR method for identifying Nosema, DNA was extracted from single normal bumblebees and bumblebees infected with microsporidia. We used genomic DNA concentrations ranging from 50 ng/μL to 0.005 ng/μL and we performed PCRs using multiple cycles (25 to 35) in order to identify the optimal concentration and cycle number for detecting Nosema. We were able identify Nosema using DNA concentration of 0.005 ng using Primer pair 1 and 2 with 35 cycles (Fig.1 and 2). However, according to a previous study (Choi et al., 2006), a nonspecific band appears when using these primers with 35 cycles, thus it is difficult to detect and verify the disease. Therefore, we repeated the PCR using a smaller number of cycles. Using Primer pair 1 (Table 2) with 25 cycles of PCR a reliable Nosema band visualized. Similarly, using Primer pair 2 (Table 2) with 30 cycles of PCR, the correct Nosema band could be detected (Figs. 3 and 4).
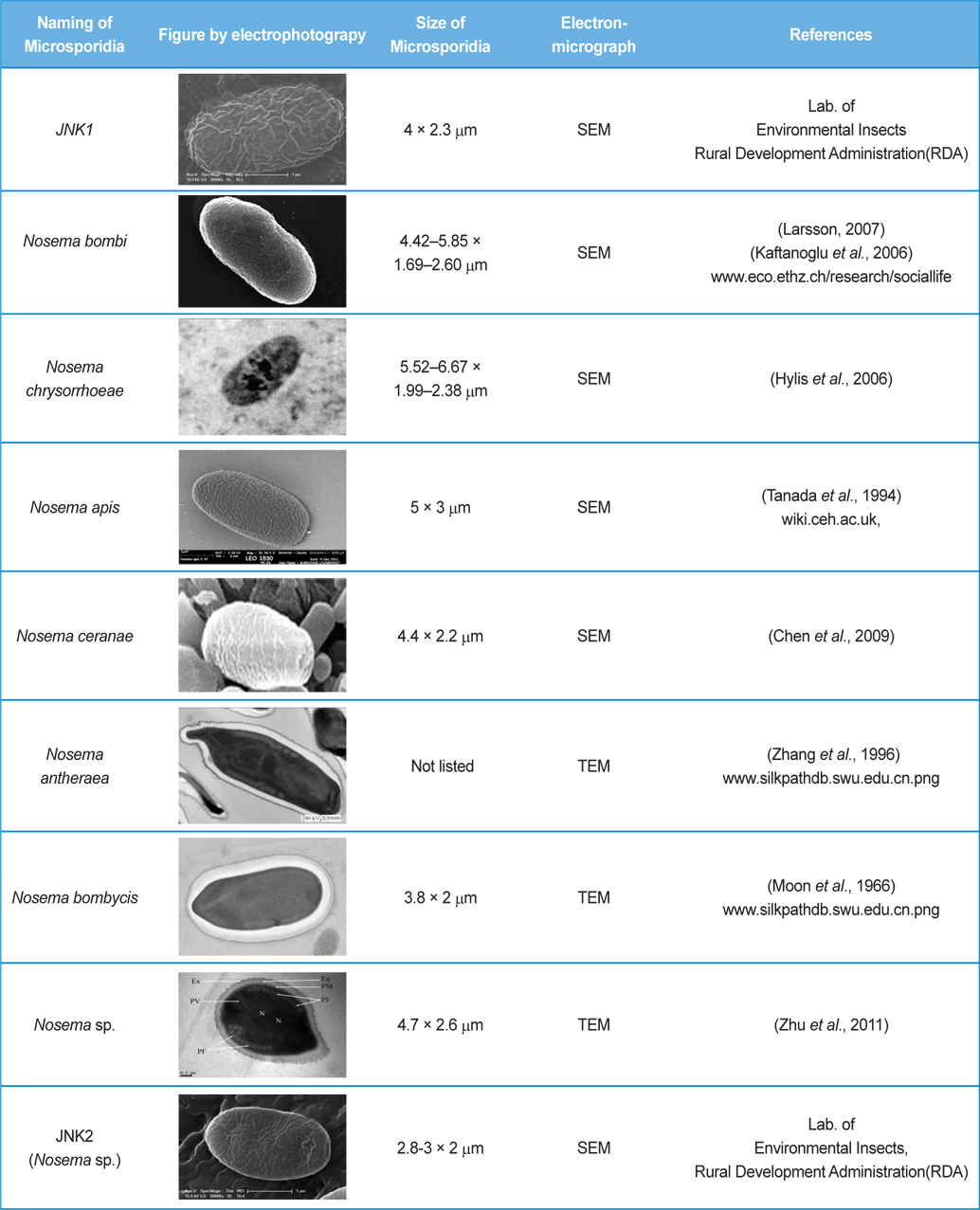
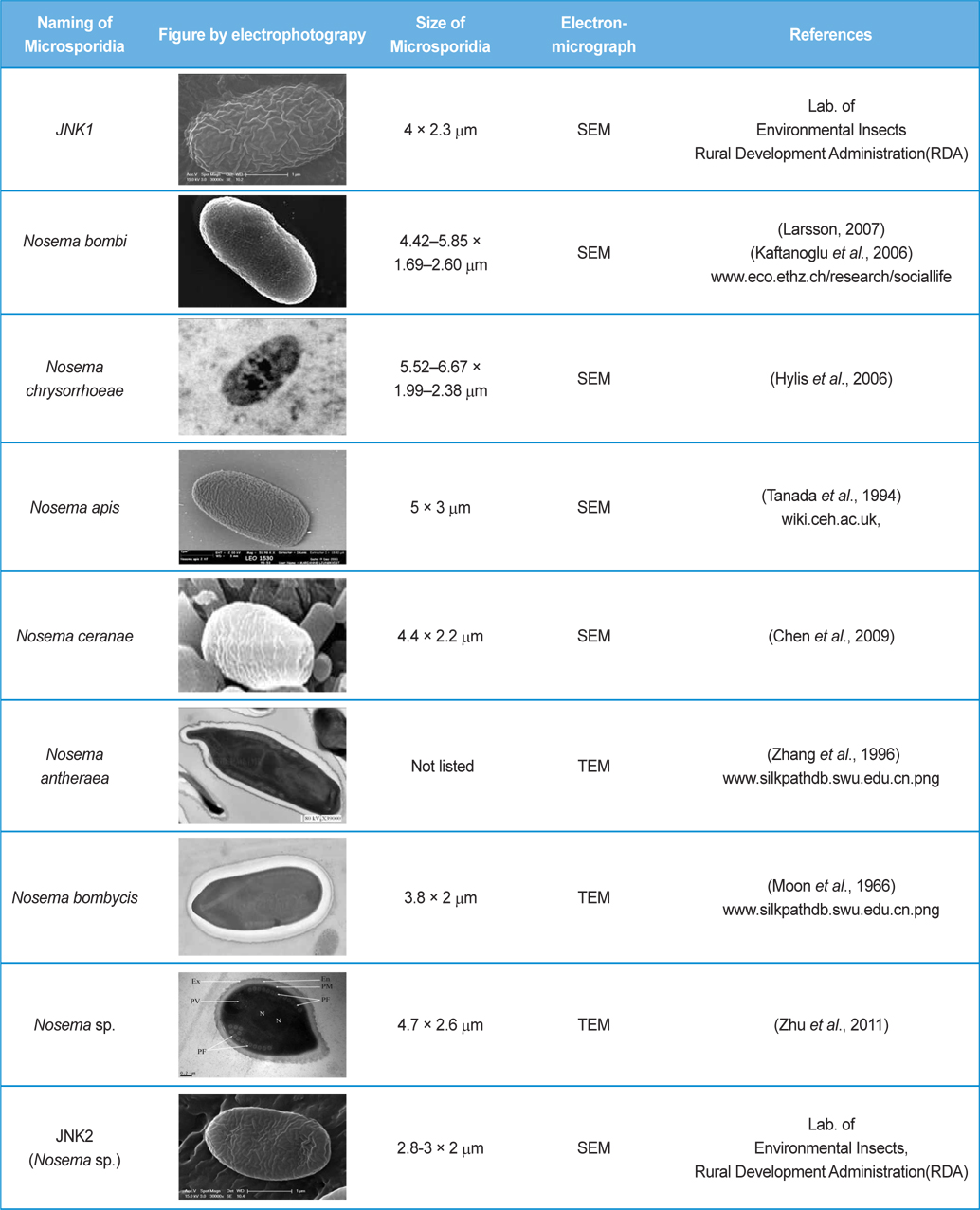
The isolated microsporidia spores were spherical and the surface structure appeared crumpled (Fig. 5). Previous studies (Tanada et al., 1994) indicate that microsporidia spore size (Table 3) is approximately 3 - 6μm × 2 - 4μm. The shapes of microsporidian spores are usually oval, ovoidal, ovocylindrical, or pyriform ; however, they are sometimes spherical, reniform, tubular, bacilliform, spiral, crescent-shaped, or comma-shaped (Tanada et al., 1994). The surface structure of microsporidia spores is used to distinguish between microsporidia species and serves as a method for classifying microsporidia (Tanada et al., 1994). The references also followed us to compare the size of N. bombi from other articles’ data (Table 3). When comparing to other figures of the Nosema species, the spore’ morphologies of N. sp., N. bombi, N. ceranae, N. apis, N. bombycis, and N. chrysorrhoeae are oval, spherical, or rod shaped. N. ceranae and N. bombi have a wrinkled and crumpled surface structure, whereas the others have almost smooth surfaces with some wrinkles. N. antheraea has curved, polar sacs not seen in Nosema species, Nosema sp. has an hooked polar sac and N. ceranae shows an inserted end part of the polar sacs. According to Fries (2010), N. ceranae has a slightly smaller spore size than N. apis ; however, the two species are difficult to distinguish under a light microscope (Fries, 2010) .
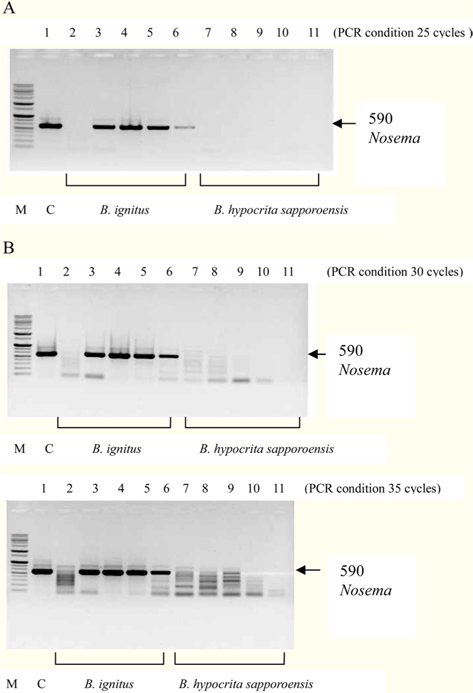
Figs. 3 and 4. Band 1: control microsporidia, Band 2: normal Bombus ignitus, Band 3:Nosema- infected queen bee B. ignitus, Band 4: Nosema-infected B. ignitus (worker bee 1), Band 5: Nosema infected B. ignitus (worker bee 2) Band 6: debris within the bumblebee hive (B. ignitus), Band 7: normal B. hypocrita sapporoensis, Band 8: B. hypocrita sapporoensis queen bee, Band 9: B. hypocrite sapporoensis (worker bee 1), Band 10: B. hypocrita sapporoensis (worker bee 2), Band 11: debris within the bumblebee hive (B. hypocrita sapporoensis). As shown in Figs. 3(C) and 4(C), the results of 35 cycle PCR conditions show the non-specific band even if the B. hypocrita sapporoensis does not have Nosema disease. Therefore, we could not verify the diagnosis of Nosema disease under 35 cycle conditions using Primer pairs 1 and 2. Primer pair 1 was used with 20 cycles (A) to check if the genomic DNA concentration could be greater than 1 ng. However, with 35 cycles condition (C), the non-specific band was seen, making it difficult to verify and diagnosis of Nosema disease by PCR. Therefore, we performed PCR under the reliable 25 cycles (B), and could identify the correct PCR condition using Primer pair 1. Therefore, we could save time and diagnose Nosema disease using Primer pair 1 with 25 cycles and Primer pair 2 with 30 cycles for PCR.
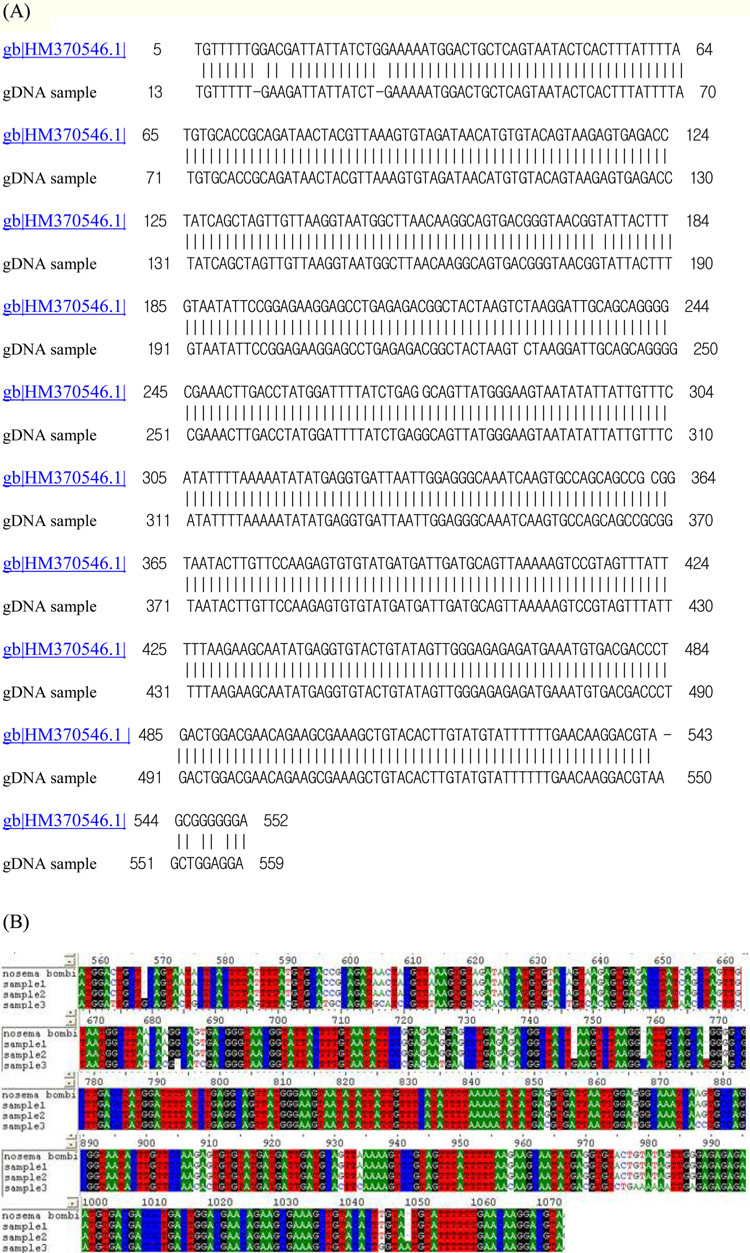
On TEM images, the internal structures of the diplokaryote of N. ceranae and N. apis were similar and N. ceranae spores contained fewer polar filament coils compared to N. apis (Fries, 2010). As JNK1 had a similar size and shape to N. bombi under SEM, we proposed that JNK1 was N. bombi, and for an accurate diagnosis, we analyzed and aligned the small subunit RNA sequence of microsporidia accordingly. The DNA sequences from JNK1 were revealed to be 99% identical to that of N. bombi (Fig. 6). According to the reports of the NCBI (Fig. 6), the identities and sequences producing significant alignment of JNK1 are N. bombi (99%), N. sp. (97%), N. thomsoni (97%), N. outlemae (96%), N. carpocapsae (96%) and N. ceranae (96%). Therefore, the reports of the NCBI verified JNK1 as N. bombi and we could identify it as such.
There are approximately 250 - 300 bumblebee species in the world, and many types of bumblebees are now being commercially mass-reared and distributed for the pollination of various crops. However, many bumblebees have become infected with pollinator parasites such as N. sp. (Graystock et al., 2013). In addition, wild bumblebee populations are known to become infected with N. bombi because of the higher infection rates seen in commercial colonies (Colla et al., 2006). Many bumblebee species are decreasing in number worldwide because of Nosema infection (Graystock et al., 2013), with many researchers suggesting that N.bombi is a fatal pathogen to bumblebees. As a microsporidian, N. bombi is a unicellular parasite that has two major life cycles, a spore stage and a vegetative stage (Otti and Schmid-Hempel, 2007). Many studies have investigated the effects of N.bombi on bumblebee health (Imhof and Schmid-Hempel, 2006), and some have reported that infected queens produce smaller colony sizes and reduced sexual offspring (Otti and Schmid-Hempel, 2008). The microsporidia are found to affect the survival and efficiency of adult individuals, as well as the sperm counts of male offspring under laboratory conditions (Otti and Schmid-Hempel, 2008). The commercialization of bumblebees as important pollinator is being actively investigated in Korea (Kim et al., 2008). In this respect, pathogens in bumblebees have to be diagnosed early so that the disease can be prevented from spreading and causing more damages to stable breeding systems. Bumblebees have excellent bee colony formations and similar genetic characteristics, conditions under which pathogens of bees could increase dramatically within colonies (Schmid-Hempel and Loosli, 1998). In this study, we sought to prompt and accurate diagnosis of Nosema from bumblebees, due to the specificity of PCR from normal B. ignitus and B. hypocrita sapporoensis being comparatively low. Under correct cycle and concentration condition of PCR, an accurate and timely diagnosis was established.
This study’s results hold particular significance as we verified the correct PCR conditions under which a diagnosis of Nosema could be made, reducing the time to diagnosis with PCR using Primer pair 1 and 2. The maximum reduction in time to diagnosis was 30 min (from 80 min to 50 min if using Primer pair 1) and the minimum detectable concentration of the sample was 0.005 ng/μL. Additionally, diagnosis of Nosema was possible by using PCR on the debris or secretions of bumblebees within the hive. As many species of wild bumblebee are infected with microsporidia, method to determine the presence or absence of microsporidia in wild bumblebees by using PCR with residues or secretions of bumblebees is of priority. PCR is a valuable tool to determine the presence or absence of Nosema disease, the early and effective detection of which will promote the breeding of healthy bumblebees and control of the disease within hives. Further, accumulation of data is required through continued investigation of the disease in bumblebees, from which a diagnosis of Nosema will be able to be made more effectively.