Oryzias latipes and Oryzias sinensis are indigenous species found in Japan, China, and other East Asian countries, including Korea. Based on morphological differences, the species have been classified distinctly. However, the range of morphological characters such as the number of gill rakers, vertebrae, and spots on the lateral body overlaps and is too vague for clear identification, so their classification based on their morphological characteristics remains uncertain. In this study, the mitochondrial cytochrome oxidase subunit I (COI) gene, which is used for DNA barcoding, was applied to clarify interspecific variation of O. latipes and O. sinensis. Intraspecific genetic diversity was calculated to identify correlations with geographic distributions. We studied two species collected from 55 locations in Korea. All individuals carried a 679-base pair gene without deletion or insertion. Between species, 525 base pairs of the gene were shared. The Kimura two parameter (K2P) distance of O. latipes and O. sinensis was 0.41% and 1.39%, respectively. Mean divergence within genera was 23.5%. Therefore, the species were clearly different. The distance between O. latipes and O. sinensis was 14.0%, which is the closest within genera. Interestingly O. latipes from the Japanese and Korean group represented 16.5% distant. These results were derived from geohistorical and anthropogenic environmental factors. The O. latipes haplotypes were joined in only one group, but O. sinensis was divided into two groups, one is found in the Han River and upper Geum River watershed; the other is found in the remaining South Korean watersheds. Further studies will address the causes for geographic speciation of O. sinensis haplotypes.
Morphological identification, the most common method for classification, has been employed for many years. Studies involve measuring and comparing the phenotypes of target species, sometimes by microscopic examination. This method is normal and sound when it applies to well-known taxa; however, it is not always efficient for species identification, unless the phenotype is obvious. To overcome this problem, a taxonomic system based on DNA analysis was established in the early 2000s (Tautz et al. 2002, 2003).
Mitochondrial DNA cytochrome oxidase subunit I (mtDNA COI) has been proposed for used in animal bioidentification because the COI gene could differentiate nearly all animal species with only a single gene sequence (Hebert et al. 2003, 2005). In the early 2000s, various taxonomic studies were carried out using the COI gene
(Hebert et al. 2003, 2004a, 2004b, 2005, Smith et al. 2006, 2007, Bucklin et al. 2010). The universal primer for the COI gene is very stable and attaches at the 5′ end of the animal gene (Folmer et al. 1994, Zhang and Hewitt 1997). The COI provides profound insights into phylogenetic studies more than other mitochondrial genes such as cytochrome b (Simmons and Weller 2001), because changes in the amino acid sequence occur more slowly (Lynch and Jarrell 1993). Moreover, mtDNA extraction and sequencing are easy because the small multi-copy mitochondrial genome is located in each cell. Inter-specific variation is high and intra-specific variation is low, making species classification clears (Jin 2012).
In this study, the mitochondrial COI region used for DNA barcoding was applied to clarify the interspecific variation of
>
Sample collection and identification of genus Oryzias
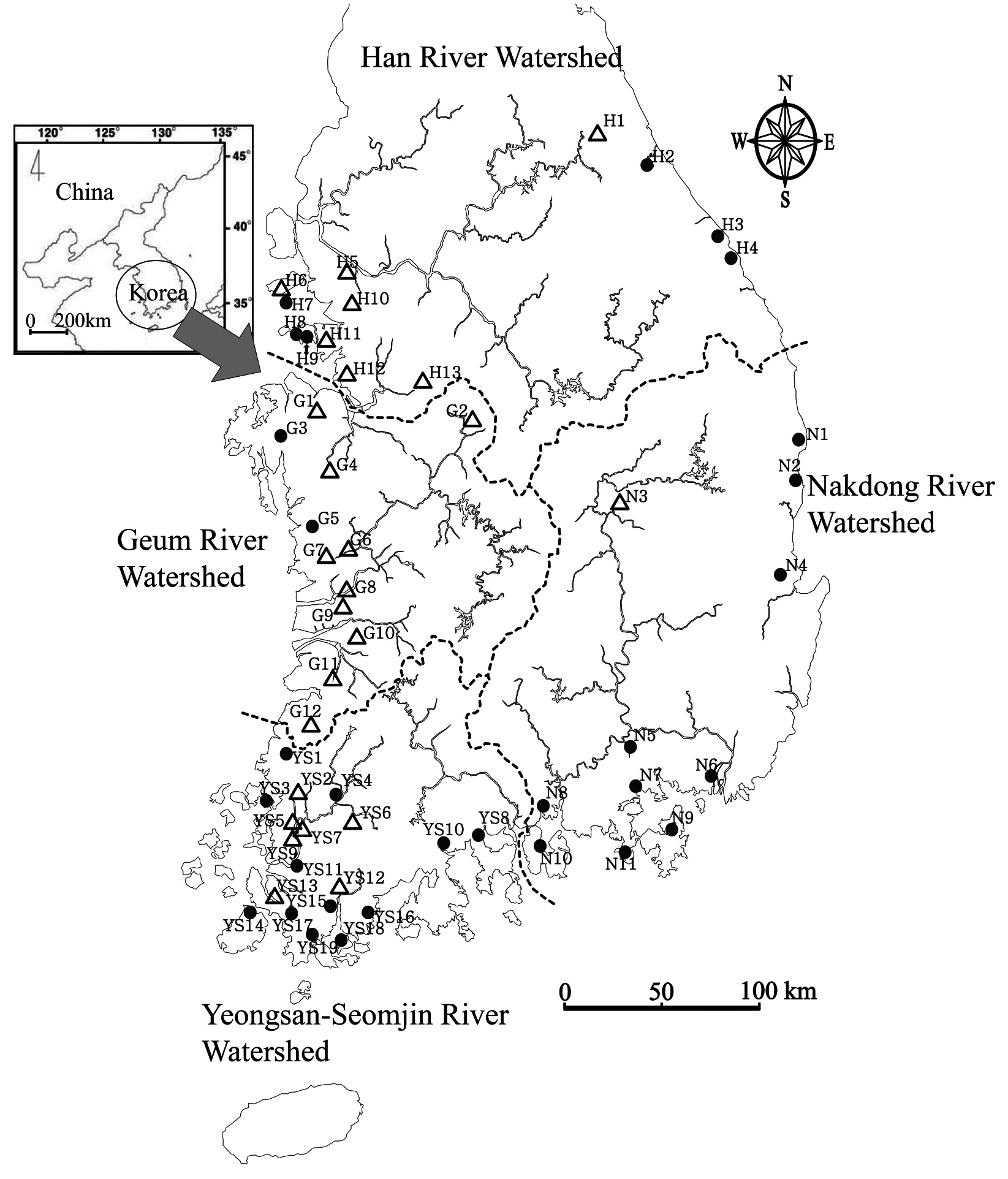
Sampling was conducted from 2009 to 2011 and sites were selected based on literature and government reports (Appendix 1). Other sites were chosen based on the possibility of
We analyzed 165 specimens from 55 sites. DNA extracts were prepared from muscle tissue (about 30 mg) using SolGent™ Genomic DNA Prep Kit (Solgent, Daejoen, South Korea). To amplify the COI gene from mitochondrial DNA, PCR was performed with primers FishF2 (5′-TCAACYAATCAYAAAGATAT-3′) and FishR2 (5′-ACTTCYGGGTGRCCRAARAA-3′) (Ward et al., 2005). The 50 μl PCR mixture included 5 μl 10× reaction buffer, 1 μl each primer (10 pmole/μl), 4 μl dNTP (2.2 mM each), 0.25 μl TaKaRa Ex Taq™ (Takara, Shiga, Japan) and 4 μl template DNA (50 ng/μl). Amplifications were performed using a C1000 gradient thermal cycler (Bio-RAD, Foster City, CA, USA). Cycling included an initial step of 3 min at 95℃ followed by 40 cycles of 0.5 min at 95℃, 0.5 min at 55℃, and 1 min at 72℃, followed by elongation for 7 min at 72℃. PCR products were analyzed by 1% agarose gel electrophoresis, and then purified with a
>
Analysis of the mtDNA COI gene
The COI sequence length was 679-bp and there were no insertions or deletions in any specimen. The COI gene in
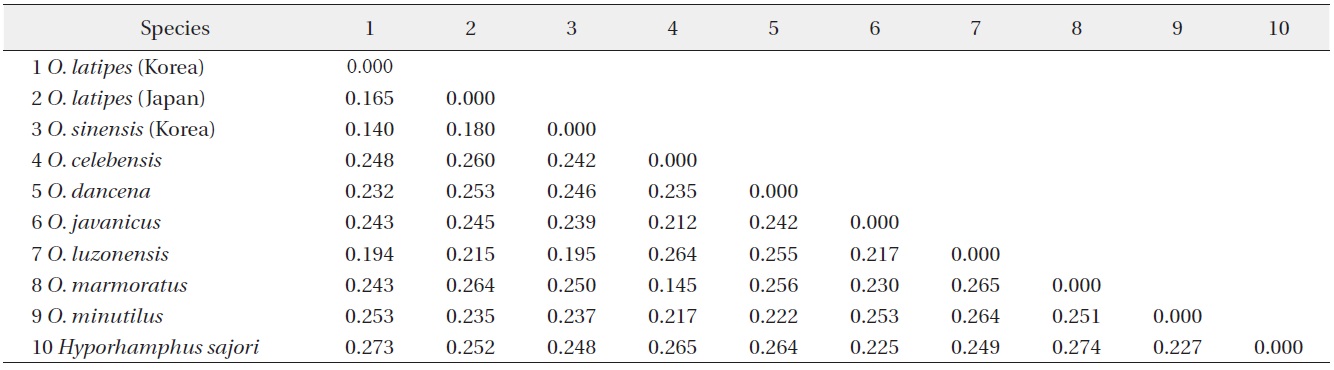
The K2P distance of
>
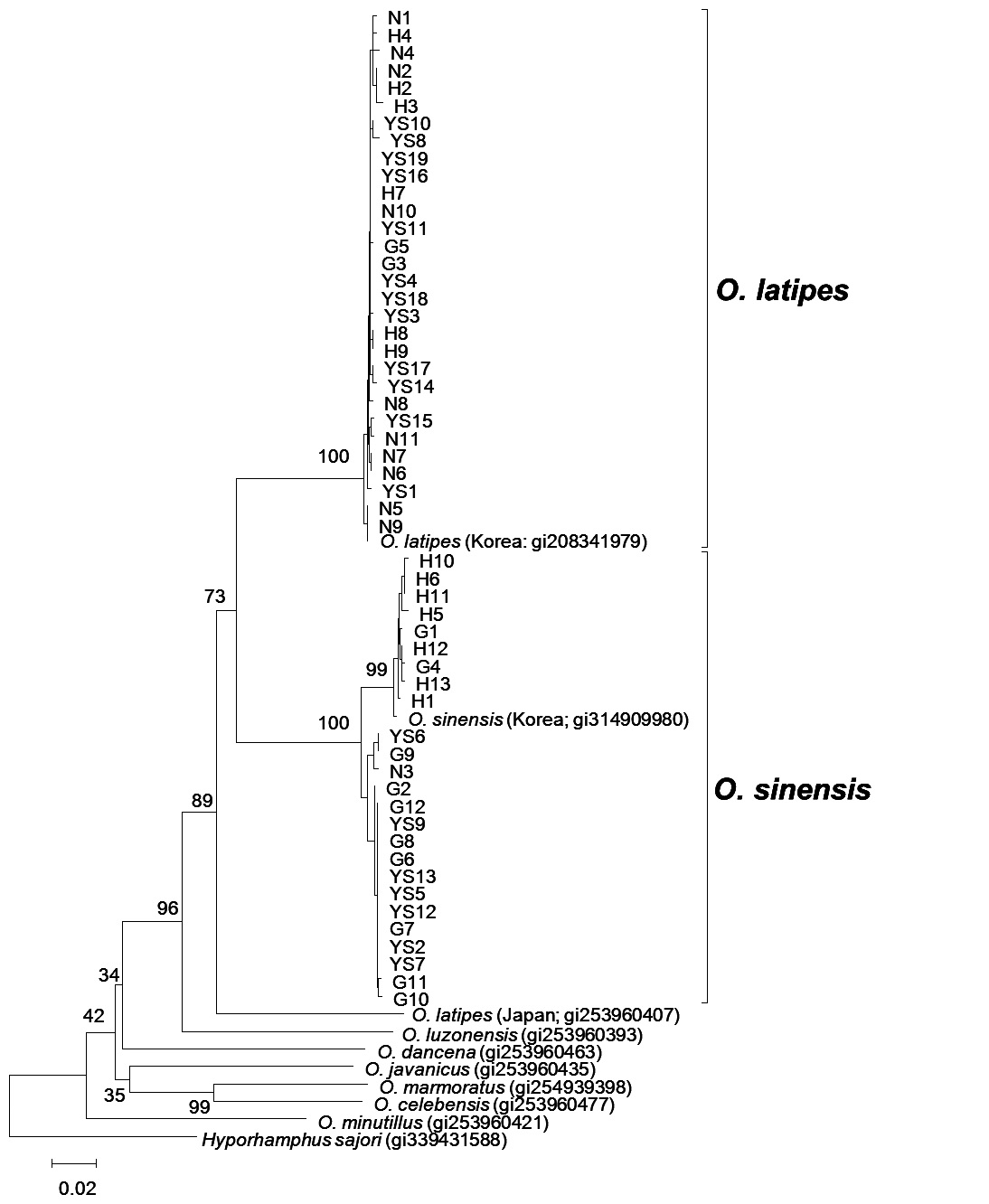
Molecular phylogeny of genus Oryzias
The molecular phylogeny results indicate
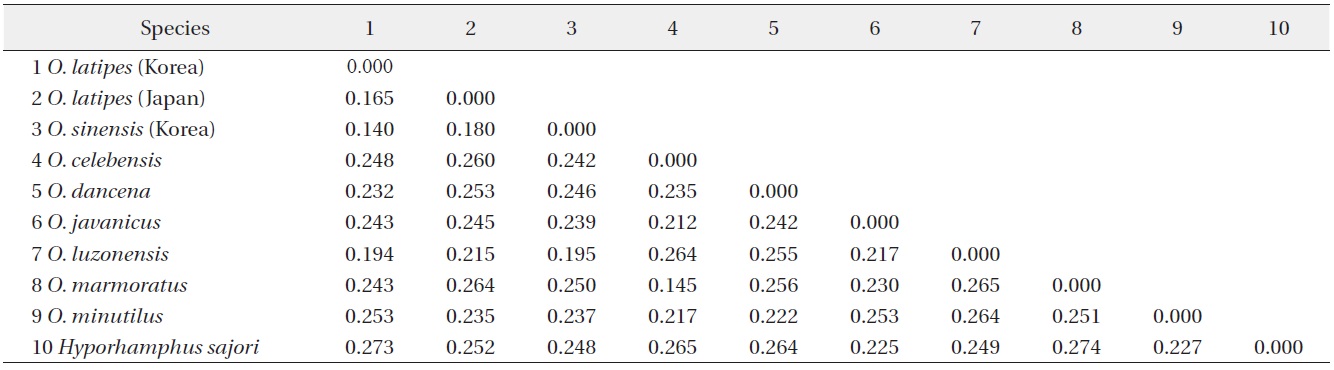
Matrix of K2P (Kimura 2 Parameter) for 9 Oryzias species. Hyporhamphus sajori is an outgroup
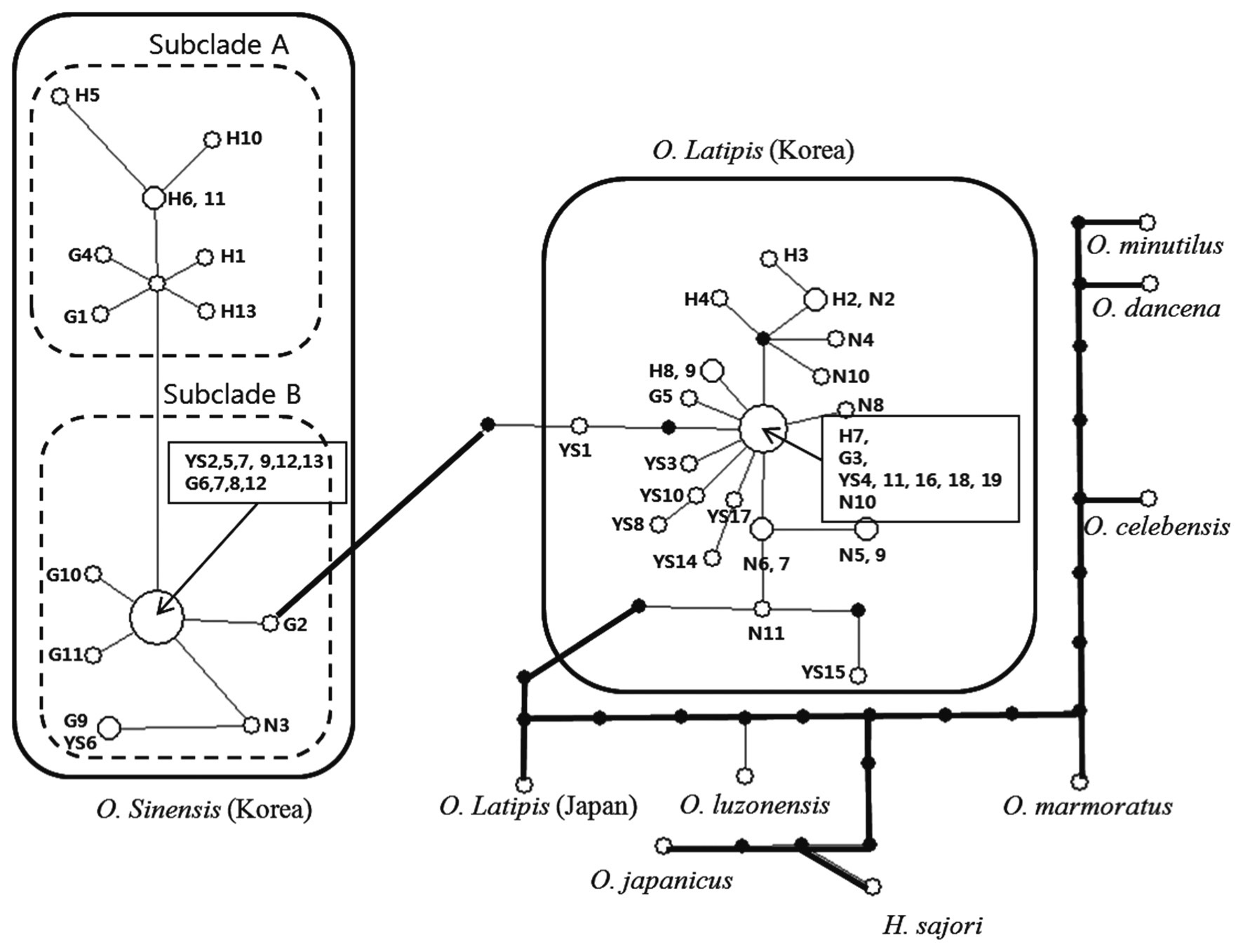
identified across 25 sites; the estimated number of mutations in the shortest tree was 32. Thus, the
>
COI region for species identification for Oryzias species
In this study, we clearly classified two species using the mitochondrial COI region. Thus, the COI region could be used to supplement current taxonomic keys that are largely focused on morphological characteristics. In many cases, morphological classification generates identification errors due to vague criteria. Therefore, species classification using DNA markers such as the COI region (DNA barcodes) is an appropriate alternative.
NJ trees clearly differentiated
>
Geographic distribution of genus Oryzias
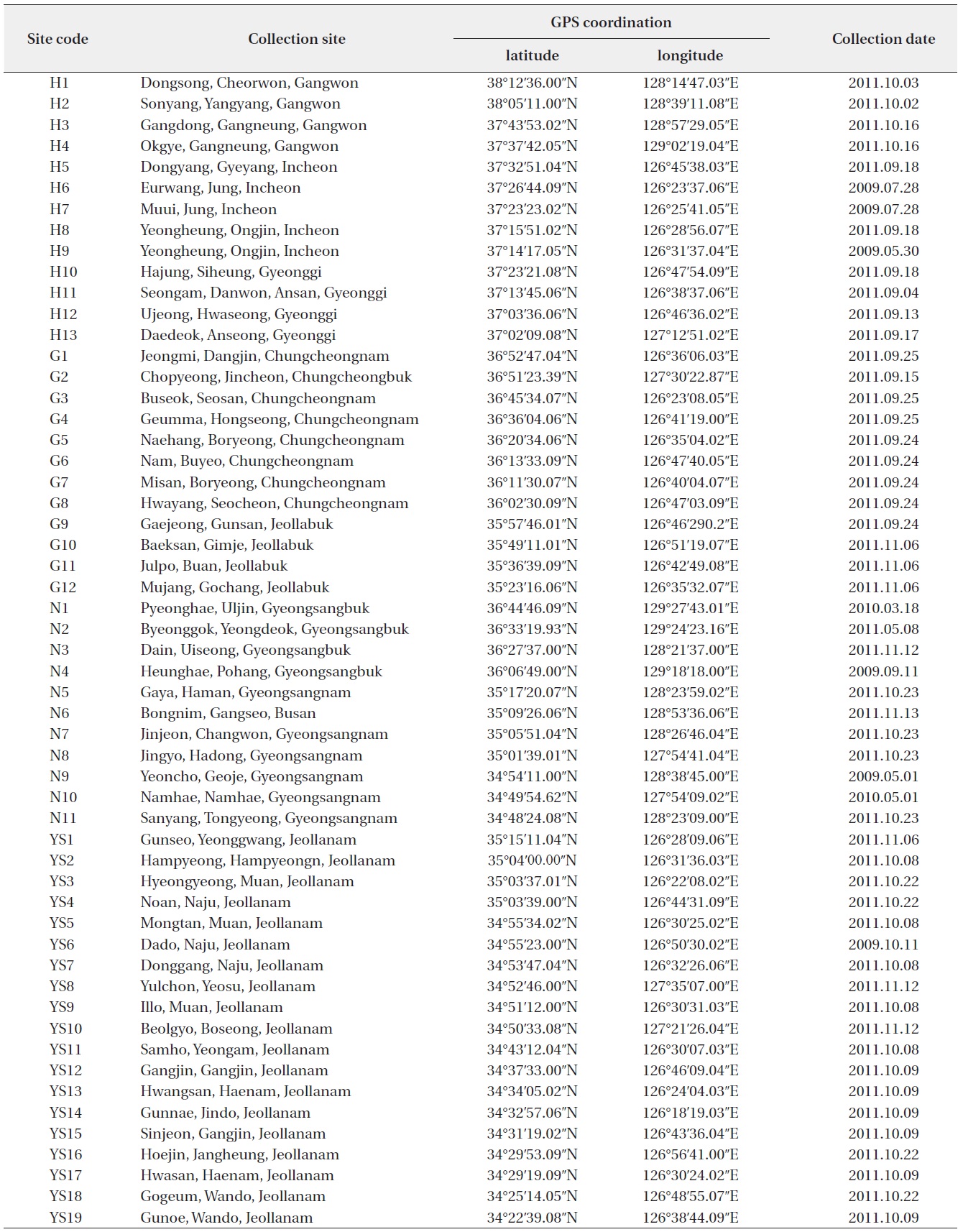
Collection sites information of genus Oryzias (H: Han River Watershed, G: Geum River Watershed, YS: Yeongsan-Seomjin River Watershed, N: Nakdong River Watershed)