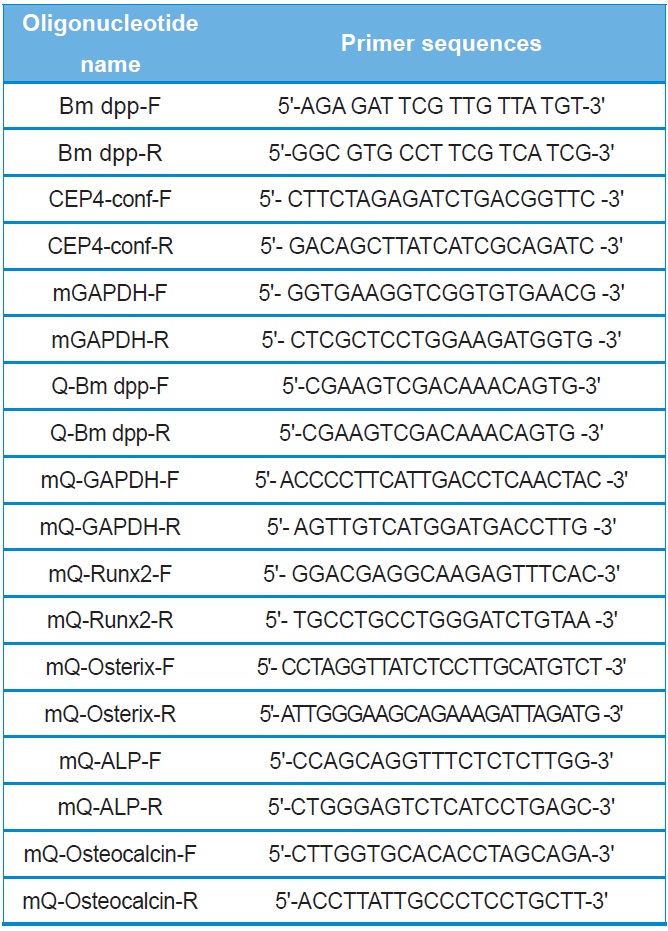
The Bombyx mori decapentaplegic (dpp) gene is conserved in the human, cow, mouse, chicken, and fruit fly genomes. In particular, the functions of dpp in Drosophila melanogaster are well known. Drosophila dpp controls multiple developmental processes and belongs to the transforming growth factor beta (TGF-β) superfamily (Chen et al., 1998; Kunnapuu and Shimmi, 2010). The TGF-β superfamily contains conserved polypeptide growth factors that play important roles in different cellular processes such as proliferation, apoptosis, differentiation and cell-fate determination (Chen et al., 1998; Kunnapuu and Shimmi, 2010). Human bone morphogenetic proteins (hBMPs) constitute the largest subgroup of the TGF-β superfamily and have the same roles as Drosophila dpp (Chen et al., 1998; Kunnapuu and Shimmi, 2010).
Osteoblast differentiation involves a complex coordination of multiple factors including several hBMPs (Aono et al., 1995; Park et al., 2012). Recombinant human BMP-2 (rhBMP-2) and BMP-4 (rhBMP-4) induce differentiation of murine mesenchymal cells to an osteogenic lineage (Bilic et al., 2006; Park et al., 2012). Interestingly, the Drosophila dpp gene is functionally interchangeable with mammalian BMP-2 and BMP-4 (Chen et al., 1998). The hBMP-4 transgene can rescue dorsal embryonic pattern defects in Drosophila dppmutant flies (Kunnapuu and Shimmi, 2010; Padgett et al., 1993). In contrast, Drosophila dpp induces bone formation in mammalian cells (Kunnapuu and Shimmi, 2010; Sampath et al., 1993). Moreover, the ligands of BMP-2, BMP-4, and dpp have evolutionarily conserved roles in embryonic development (Kunnapuu and Shimmi, 2010).
The B. mori dpp gene shares genetic homology with hBMPs and Drosophila dpp. Few studies have been conducted to examine the functions of B. mori dpp; therefore, its function is not well understood. In this study, we showed that B. mori dpp is expressed in C3H10T1/2 pluripotent stem cells (bi-potential mesenchymal precursor cells) transfected with pCEP4-Bm dpp plasmid DNA. These cells exhibited biological activity consistent with the roles of dpp as an osteogenic differentiation induction growth factor, similar to hBMPs. We performed quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) to evaluate B. mori dpp-induced RUNX2, osterix, osteocalcin, and alkaline phosphatase enzyme (ALP) expression. Our results showed that B. mori dpp is functionally interchangeable with hBMP-2, hBMP-4, and Drosophila dpp and can be used in tissue engineering of bone tissue substitutes.
All recombinant DNA manipulations were performed using standard techniques (Russell and Sambrook, 2000). Total RNAs were isolated from the fat body of 3-day-old of the 5th instar larvae using the TRIzol reagent according to the manufacturer's instructions (Invitrogen). Amounts of total RNAs were determined spectrophotometrically by measuring the absorbance at 260 nm. RNAs were stored at ?70℃ until use. After purification, oligo dT-primed cDNAs were prepared from 2 μg of total RNAs using the High-Capacity cDNA Archive kit (Applied Biosystems). The reaction was allowed to proceed for 2 hours at 37℃. The 1,285-bp full-length cDNA of B. mori dpp was amplified using primers (forward: Bm dpp-F and reverse:
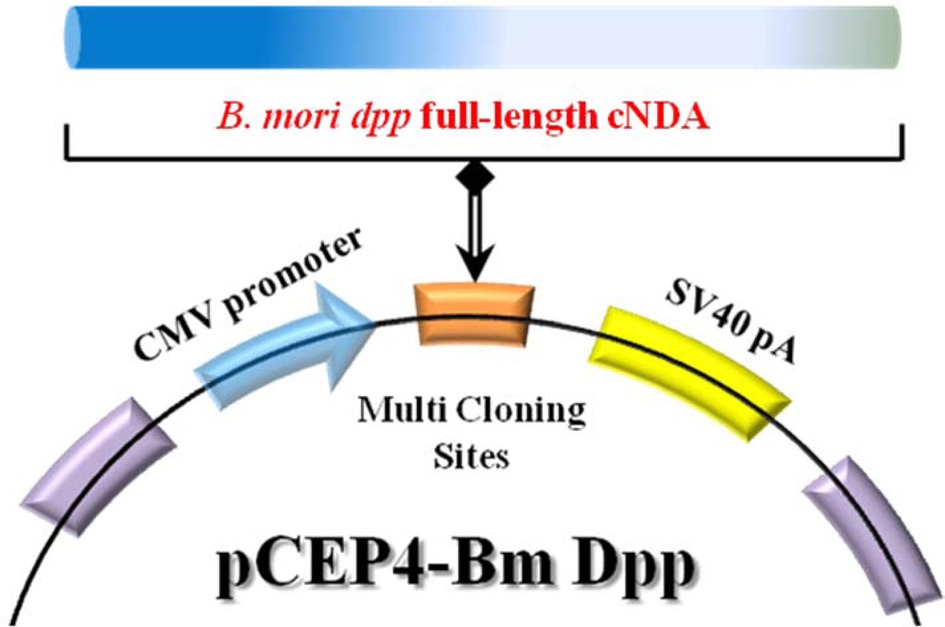
Bm dpp-R). The primer pairs are shown in Table 1. Top-Taq PreMix (CoreBio Systems) polymerase was used for cDNA amplification. The annealing temperature was 55℃, and the PCR reaction was conducted for 35 cycles. The PCR fragment was cloned into the pGEMT-Easy vector (Promega). The resulting plasmid, reffered to as pSRDA-1, was used for DNA sequencing (CoreBio Systems). Sequence data of the DNA fragments were analyzed by using the Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov). To generate the B. mori dpp expression vector, the dpp gene was excised as a 1,324-bp fragment following NotI restriction enzyme digestion from the pSRDA-1 plasmid DNA. This fragment was inserted into the NotI restriction site of the pCEP4 mammalian expression vector (Invitrogen) in a direct orientation with respect to the Cytomegalovirus (CMV) promoter to generate pCEP4-Bm dpp (Fig. 1).
C3H10T1/2 cells were a gift from Dr. D. C. Kang from the Ilsong Institute of Life Science, Hallym University and grown in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum. Cells were plated in a 12-well tissue culture plate (1×103 cells per well). After 24 hours, the medium was replaced with 1.0 mL of fresh serum-free medium. After additional incubation for 1 hour, the cells were transfected with 1.0 μg of pCEP4 vector or pCEP4-Bm dpp plasmid DNA using the Fugene Transfection Reagent (Roche). Four hours after transfecion, the cells were washed twice, and 1.0 mL of medium was added. At 48 hours post-transfection, the medium was removed, and 1.0 mL of fresh medium containing hygromycin B (HygB; 500 μg/mL) was added. After 10 days, HygB resistant cells were designated as B. mori dpp-expressing cells.
Two PCR primers were used to confirm that the pCEP4 vector or pCEP4-Bm dpp plasmid DNAs had been transfected into the cells (forward: CEP4-conf-F and reverse: CEP4-conf-R). Genomic DNA was isolated from selected cell clones using the TRIzol reagent. A 5-μL aliquot of genomic DNA from each sample was mixed with Taq DNA polymerase in PCR buffer and amplified for 35 cycles at an annealing temperature of 58℃. To detect the specific expression patterns of the B. mori dpp gene in the HygB-resistant cells, total RNAs were treated with DNase I for 1 hour at 37℃ to remove genomic DNA. After purification, oligo dT-primed cDNAs were synthesized from 5 μg of total RNA using the High-Capacity cDNA Archive kit. The reaction was allowed to proceed for 2 hours at 37℃. Specific primers were used for RT-PCR (forward: Q-Bm dpp-F and reverse: Q-Bm dpp-R). The PCR reaction was conducted for 35 cycles at an annealing temperature of 58℃. PCR products were analyzed using 2.0% agarose gel.
We conducted quantitative RT-PCR in a 20-μL system containing 10-μL of SYBR Premix Ex Taq according to the manufacturer’s instructions (TakaRa). To quantify the amount of RUNX2, osterix, osteocalcin, and ALP expression, an endogenous internal control gene [mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH)] was used as an internal reference to normalize the quality of total RNAs purified from 3 different cell types (C3H10T1/2 cells, C3H10T1/2 cells transfected with pCEP4, and C3H10T1/2 cells transfected with pCEP4-Bm dpp plasmid DNA). Real-time PCR
was performed using the ABI7300 real-time PCR instrument (Applied Biosystems). Fold changes in gene expression were determined using the comparative CT method as described in the ABI Prism 7700 Sequence Detection System User Bulletin #2 (Applied Biosystems).
The 1,285-bp full-length cDNA of B. mori dpp was amplified and cloned into the pGEMT-Easy vector. Sequence data from the analysis of B. mori dpp full-length cDNA was compared with known sequences (http://www.ncbi.nlm.nih.gov/nuccore/FJ572058); in total, 7 base pairs were found to be replaced (Fig. 2). Next, to produce a B. mori dpp-expressing plasmid that could induce osteogenic differentiation, full-length B. mori dpp cDNA sequences were inserted in the pCEP4 mammalian expression vector. This gene is commonly under the control of the CMV promoter, so expression occurs in all mammalian cells. The detailed procedure for construction of recombinant pCEP4-Bm dpp plasmid is shown in Fig. 1.
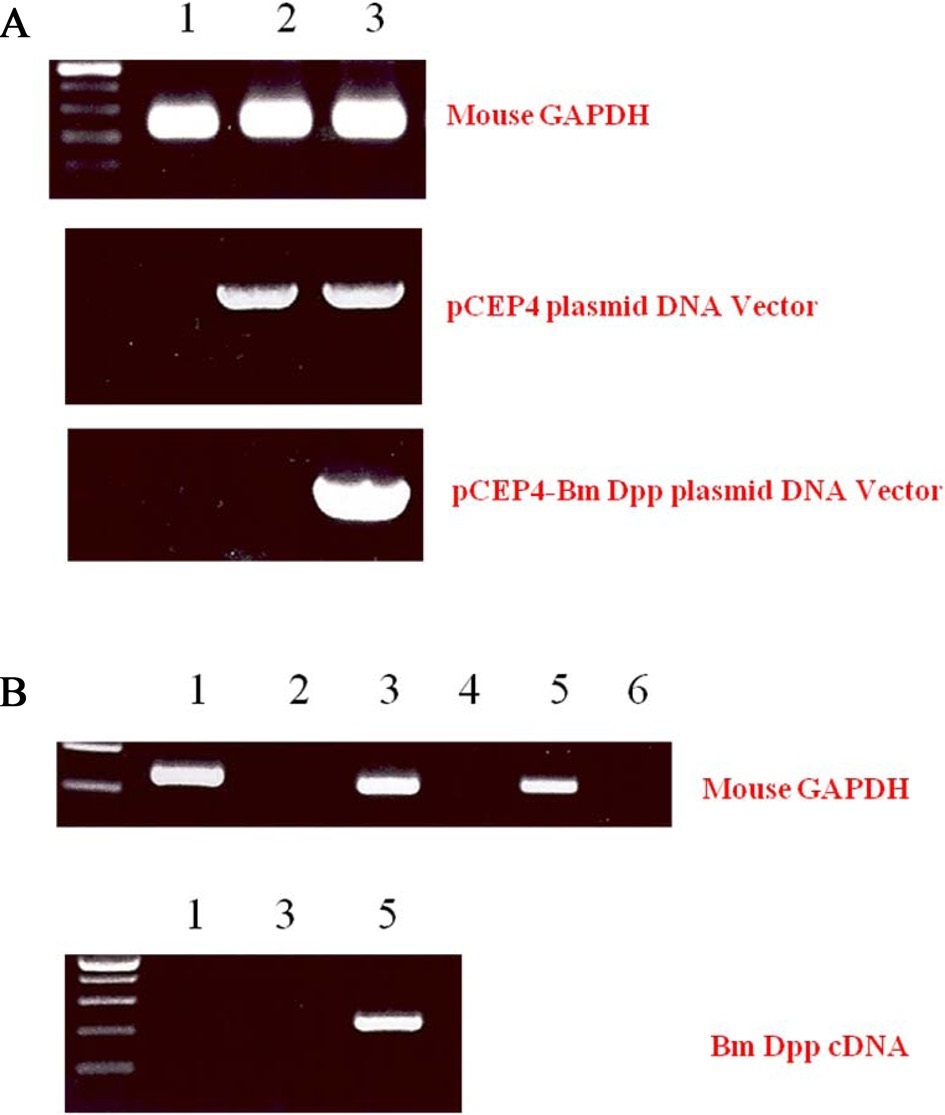
To remove as many control cells as possible, C3H10T1/2 cells transfected with pCEP4 or pCEP4-Bm dpp plasmid DNAs were cultured with the antibiotic reagent HygB for 10 days. Expression of B. mori dpp gene transcripts in the HygB-resistant cells was examined by performing genomic DNA PCR. As shown in Fig. 3A, an 835-bp fragment was amplified from the genomic DNA of cells transfected with the pCEP4 or pCEP4-Bm dpp plasmid DNAs. A 213-bp fragment of the B. mori dpp gene was amplified from only 1 sample that was transfected with the pCEP4-Bm dpp plasmid DNA. These results suggest that almost HygB-resistant cells contained pCEP4 or pCEP4- Bm dpp. RT-PCR analysis was performed to determine whether the HygB-resistant cells transfected with the pCEP4-Bm dpp plasmid expressed recombinant B. mori dpp protein. We performed 35 cycles of PCR to amplify RNA transcripts derived from control, HygB-resistant cells transfected with the pCEP4, and HygB-resistant cells transfected with the pCEP4-Bm dpp plasmid. As shown in Fig. 3B, moue GAPDH was detected in all samples. However, a 213-bp fragment was amplified from an RNA transcript obtained from HygB-resistant cells transfected with the pCEP4-Bm dpp but not from the RNA transcripts
obtained from the other 2 cell types. This result shows that the target cells expressed the B. mori dpp gene, and could be used to examine induction of osteogenic differentiation.
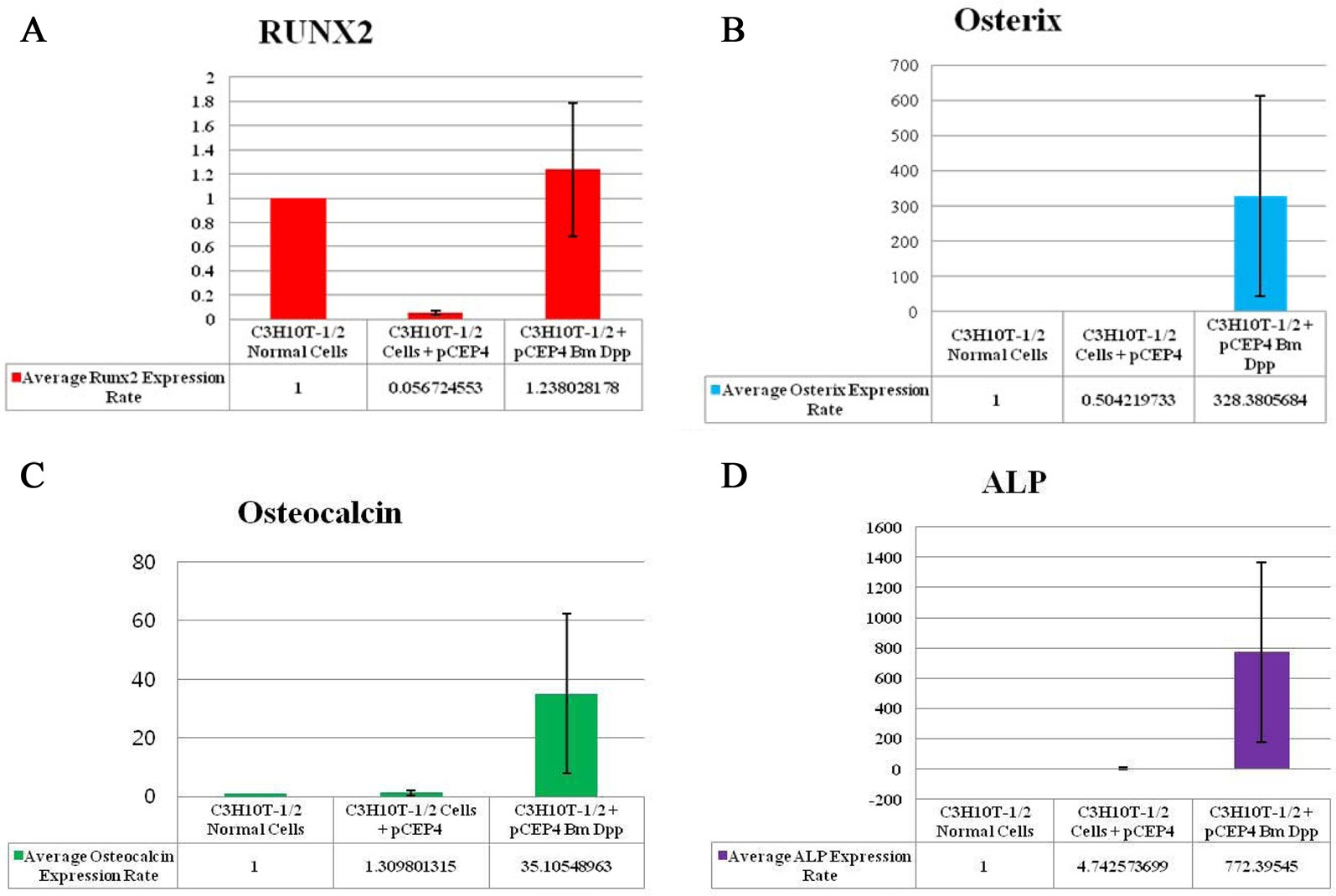
To verify the osteoblast-specific differentiation of the target cells, we performed qRT-PCR. Expressions of 4 osteogenic differentiation marker genes (RUNX2, osterix, osteocalcin, and ALP) were normalized to the expression levels of the endogenous mouse GAPDH gene as an internal reference. The mRNA expression rate of each osteogenic differentiation marker gene isolated from target cells was compared with that in control cells and HygB-resistant cells transfected with the pCEP4 plasmid. CT values for mRNA expression of RUNX2, osterix, osteocalcin, and ALP, indicated their expression in the bone-specific markers that were examined. We observed that the expression rates of 3 of the marker genes (osterix, osteocalcin, and ALP), but not RUNX2, in target cells were much higher than those in control cells and HygB-resistant cells transfected with the pCEP4 plasmid (Fig. 4). These data indicate that the B. mori dpp protein was in the target cells, and this protein switched the differentiation pathway of the target cells into that of osteoblast lineage cells.
Osteoblast differentiation involves a complex coordination of multiple factors including several of the BMPs (Sangadala et al., 2009). Previous studies have reported that the bone-forming activity of the Drosophila dpp protein can be demonstrated by measuring the specific activity of ALP and the calcium content in subcutaneous rat implants (Sampath et al., 1993). The B. mori dpp gene shares genetic homology with hBMPs and Drosophila dpp. However, functional analyses of this gene have not been conducted, so far. To investigate the biological roles of the B. mori dpp gene, we constructed a pCEP4-Bm dpp expression cassette vector under the control of the CMV promoter for producing a recombinant protein in the mammalian pluripotent stem cells. C3H10T1 /2 cells transfected with this plasmid produced the B. mori dpp protein. Furthermore, the target cells were differentiated to osteoblast lineage cells. This finding demonstrates that the B. mori dpp gene is functionally interchangeable with mammalian BMPs and Drosophila dpp.
RUNX2 is a key transcription factor in the differentiation of mesenchymal precursors to osteoblasts (Matsumoto et al., 2010). Firstly, BMP treatment significantly increases mRNA levels of RUNX2. Secondly, RUNX2 directly regulates expression of osteocalcin (Matsubara et al., 2008). BMP2 is known to control the expression and functions of RUNX2 through Smad signaling, and a BMP2-Smad-RUNX2 axis in osteoblastogenesis has been established (Matsubara et al., 2008). However, our study showed that RUNX2 transcription levels in the B. mori dpp-expressing C3H10T1/2 target cells were not different from those in control cells and C3H10T1/2 cells transfected with the pCEP4 (Fig. 4A). Thus, the B. mori dpp directly regulates osterix expression and does not affect the BMP2-Smad-RUNX2 axis (Fig. 4B). Osterix is also up-regulated by BMP2 during osteoblastic differentiation (Matsubara et al., 2008; Nakashima et al., 2002). Osterix is thought to function downstream of RUNX2 during osteoblast differentiation (Choi et al., 2011; Matsubara et al., 2008). Recently, Matsubara et al.(2008)
reported that osterix expression is regulated via both RUNX2-dependent and -independent mechanisms via BMP2 signaling. Therefore, this study shows novel biological effects of B. mori dpp in osteoblastic differentiation. The B. mori dpp may control C3H10T1/2 cell osteogenesis by regulating target genes downstream of RUNX2.
Our results demonstrate the role of the B. mori dpp in increasing the levels of osteogenic-specific genes in mouse pluripotent stem cells during osteogenic differentiation. Recombinant BMPs can be delivered via the culture medium to regulate cellular processes in the field of tissue engineering. However, the short half-lives, the requirement of relatively high levels, high costs, and potential toxicities at the systemic level have hindered many applications for these bioactive compounds (Edelman et al., 1993; Wang et al., 2009). Further studies examining the B. mori dpp should be conducted to create stable cell lines, such as bio-medical insect cells, that can be used to produce bone growth formation biomaterials. The recombinant B. mori dpp protein will be functionally and economically useful for tissue engineering of bone tissue substitutes.