Zygnema J. Agardh (1824) is a conjugating unbranched filamentous green algal genus that is widely distributed in aquatic habitats, from sea level to alpine summits (Transeau 1951). The slimy masses of this alga, less slippery than the Spirogyras but more slippery than the Mougeotias, occur in small lotic or lentic bodies of water (Transeau 1951, Bold and Wynne 1985). However, their life cycle is completed in a few weeks, and reproductions are frequently found in temporary ponds and ditches (Transeau 1951). Non-flagellated amoeboid gametes, sexual reproduction by isogametes, and short cylindrical cells containing a pair of stellate chloroplasts are features that assign Zygnema to the family Zygnemataceae (Smith 1933, Transeau 1934).
Since the description of Zygnema by J. Agardh (1824), more than 150 species have been described by taxonomic studies (Randhawa 1959, Kadlubowska 1984, Rundina 1998, Novis 2004, Zarina et al. 2006). Morphological criteria such as vegetative cell size, details of sexual reproduction, shape, dimension and color of zygospores, and ornamentation of the median spore wall are important in the identification of species (Randhawa 1959, Kadlubowska 1984, Johnson 2002). However, features such as cell width, and size of the zygote are highly variable in both culture and nature. For example, the variation of cell width in Z. circumcarinatum clones was much higher than expected (Miller and Hoshaw 1974), and these ploidal variants in clonal culture and field collected materials have led to a proliferation of species diversity in Zygnemataceae (McCourt and Hoshaw 1990).
Molecular studies of Zygnema began with phylogeny of the family Zygnemataceae, in which plastid rbcL and nuclear small subunit of ribosomal gene (SSU rDNA) were analyzed for a few species (McCourt et al. 1995, 2000, Gontcharov et al. 2003, 2004, Hall et al. 2008). Recently, Stancheva et al. (2012) gave an intensive phylogeny of Zygnema from California, based on the cox3 and rbcL genes, recognizing two clades within the genus. However, the relationships within Zygnema remain unresolved due to limited taxon sampling.
To date, eleven species of Zygnema have been recorded by floristic studies on local populations in Korea (Chung 1968, 1970, Wui and Kim 1990, Kim and Kim 2009): Z. carinthiacum Beck, Z. cruciatum (Vaucher) Agardh, Z. decussatum (Vaucher) Agardh, Z. insigne (Hassall) Kutzing, Z. leiospermum De Bary, Z. pectinatum (Vaucher) Agardh, Z. peliosporum Wittrock, Z. shigaense Yamagishi, Z. stellinum (Vaucher) Agardh, Z. sterile Transeau and Z. vauche-
rii Agardh. Of these, Z. cruciatum is the only species for which the morphology was studied by light microscopy and scanning electron microscopy (Kim and Kim 2009). There are no phylogenetic studies of Korean Zygnema probably owing to difficulties in their identification and limited sampling of fertile specimens.
In the present study, we investigate the morphology and psbA (encoding the photosystem II thylakoid protein D1) sequences of two Zygnema species, Z. insigne and Z. leiospermum, in Korea, with the aim of clarifying their taxonomic identities. One species of Zygnema from the Culture Collection of Algae and Protozoa (CCAP) was included for a better understanding of phylogeny. Putative relatives, Spirogyra and Mougeotia, were included in the study as outgroup. This is the first report on the psbA phylogeny of Zygnema.
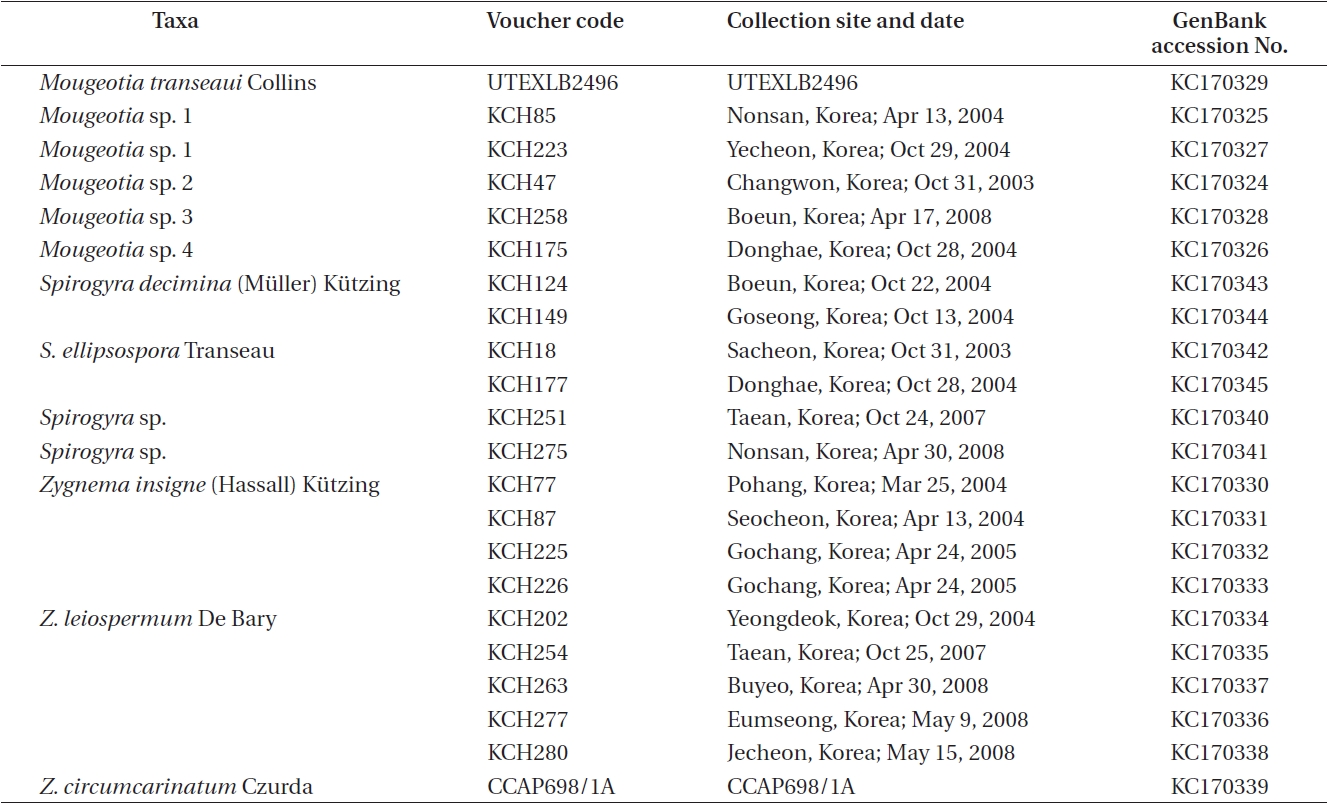
A total of 22 isolates of Zygnema and putative relatives were included in the present study (Table 1). The strains of nine Zygnema were isolated from various freshwaters bodies across South Korea, and vegetative filaments containing 2-3 cells of each collection were used for unialgal culture. For the present study, six Spirogyra and five Mougeotia isolates from Korea were also included. In addition, one Zygnema and one Mougeotia isolate were obtained from CCAP and the Culture Collection of Algae at the University of Texas at Austin (UTEX), respectively. All isolates were grown in Wood Hole liquid medium buffered to pH 7.0 (Nichols 1973). Cultures were maintained at 20 ± 1℃ on a 16 : 8-h light : dark cycle under 30-50 illumination with μmol m-2 s-1 with cool-white fluorescent lamps (Pringsheim 1967, Stein 1973). Morphological features were observed under a light microscope (Nikon Optiphot; Nikon, Tokyo, Japan) equipped with the Nikon UFX-II camera. Voucher specimens were deposited at the herbarium of Chungbuk National University (CBNU), Cheongju, Korea.
Live or air dried specimens from unialgal cultures of each strain were used for DNA extraction. Genomic DNA was extracted from approximately 0.01 g of algal powder, ground in liquid nitrogen, using the DNeasy Plant Mini Kit (Qiagen Gmbh, Hilden, Germany). Extracted DNA was dissolved in 150 μL of distilled water, stored at -20℃ and was used to amplify the psbA gene.
The psbA region was amplified with psbA-F and psbAR2 primers as described by Yoon et al. (2002). PCRs were carried out in a 25 μL reaction volume containing 10× Ex Taq buffer, 25 mM MgCl2, 2.5 mM of each dNTP, 10 pmole of each primer, 5 U Taq polymerase (Takara Ex Taq; Takara Bio Inc., Tokyo, Japan), 25-50 ng DNA template, and distilled water. Amplification was performed using a modified protocol of McCourt et al. (1995). The PCR started with an initial denaturation cycle at 95℃ for 4 min, followed by 33 cycles of denaturation at 95℃ for 1 min, primer annealing at 47℃ for 1 min, and an extension at 72℃ for 2 min. The amplification was terminated with a final extension at 72℃ for 6 min. PCR products were purified using the High Pure PCR Product Purification Kit (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer’s instructions. The sequences of the forward and reverse strands were determined for all strains using an ABI PRISM 377 DNA Sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA, USA). The electropherogram output for each specimen was edited using the program Sequence Navigator v. 1.0.1 (Applied Biosystems).
Twenty-three psbA sequences, including previously published sequence of Zygnema circumcarinatum SAG698-1a (Turmel et al. 2005), consisting of eleven Zygnema, six Spirogyra, and six Mougeotia, were collated using the multisequence editing program, SeqPup (Gilbert 1995) and were aligned by eye. There were no gaps in our alignments of the psbA region.
Phylogenetic trees were reconstructed using maximum parsimony (MP), maximum likelihood (ML) and Bayesian analyses (BA). MP trees were constructed with PAUP* 4.0b.10 (Swofford 2002) using a heuristic search algorithm with the following settings: 100 random sequence-addition replicates, tree bisection-reconnection (TBR) branch swapping, MulTrees, all characters unordered and unweighted, and branches with a maximum length of zero collapsed to yield polytomies. Bootstrap values (BS) for nodes in the tree were obtained from 1,000 bootstrapping replicates (Swofford 2002).
For ML and BA, a likelihood ratio test was carried out with ModelTest 3.08b (Posada and Crandall 1998). The general time reversible (GTR) model, with a gamma correction for rate variation across sites (G) and proportion of invariable sites (I), was chosen as the best model for our data. ML analysis was conducted using PAUP* with GTR + I + G model. To find the best tree, we used a heuristic search with 100 random sequence-addition replicates, TBR branch swapping, and MulTrees options. The ML BS for each branch was estimated by performing 1,000 replicate ML searches, with two random addition sequence replicates.
BA were performed using MrBayes 3.0 (Huelsenbeck and Ronquist 2001). Each analysis was initiated from a random starting tree, and the program was set to run four chains of Markov chain Monte Carlo iterations simultaneously for 1,000,000 generations, with tree sampling every 100th generation. The likelihood scores stabilized at approximately 110,000 generations, and thus the first 1,100 trees were burned.
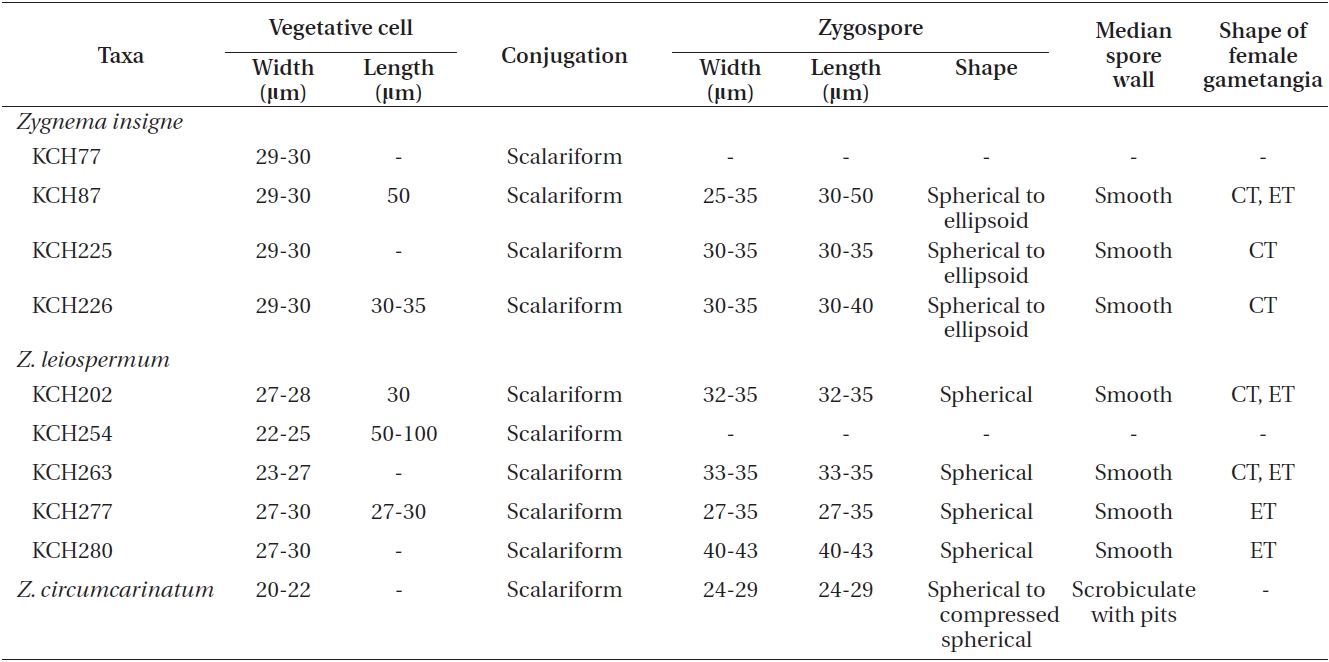
We collected nine isolates of Zygnema insigne and Z. leiospermum from Korean water systems. All isolates were cultured in the laboratory and the cultured materials were compared with field-collected specimens. However, most of quantitative characters were based on the field specimens. A comparative morphology of two species is given in Table 2.
Kutzing 1849, p. 444; Czurda 1932, p. 127, Fig. 131; Jao 1935, p. 567, Pl. 1, Fig. 6; Transeau 1951, p. 35; Randhawa 1959, p. 234, Fig. 176; Kadlubowska 1984, p. 201, Fig. 297.
Basionym. Tyndaridea insignis Hassall 1843.
Specimens examined. Rice field, Pohang, Korea (Mar 25, 2004); Ditch water, Seocheon, Korea (Apr 13, 2004); Ditch water, Gochang, Korea (Apr 24, 2005); Rice field, Gochang, Korea (Apr 24, 2005).
World distribution. Australia, China, Europe, India, Japan, Pakistan, and the United States of America.
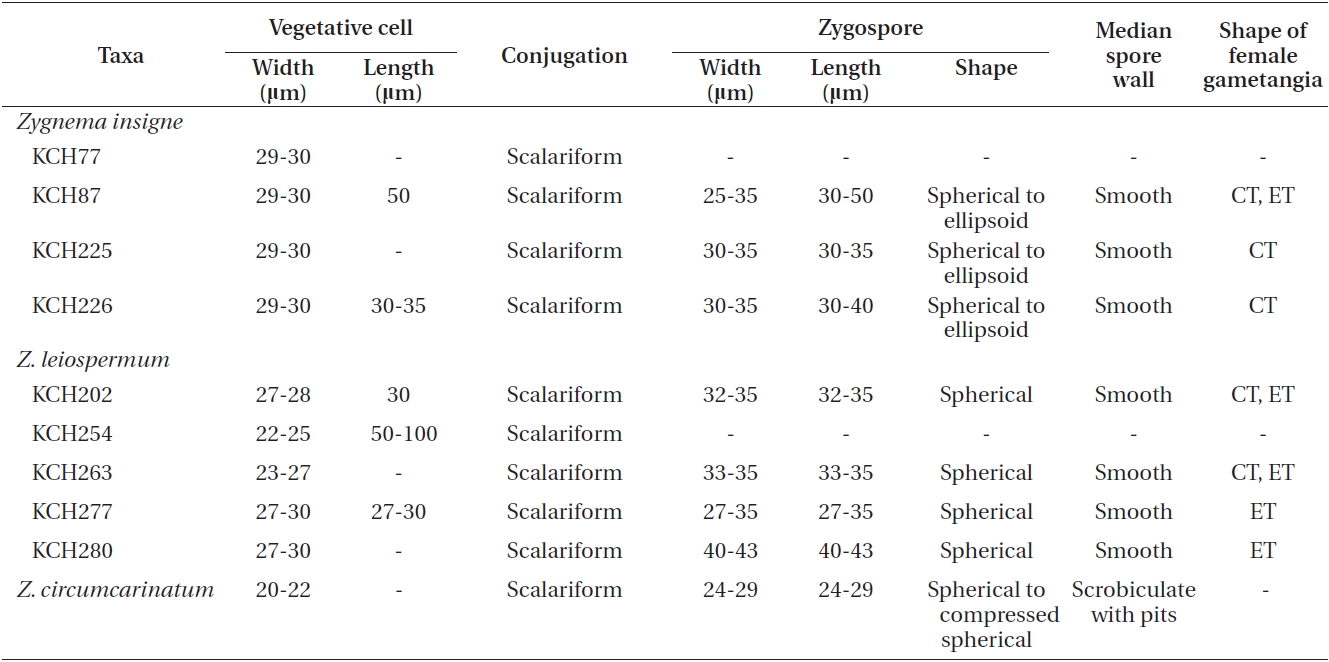
Morphology. Plants are unbranched filamentous of short cylindrical cells with plane end wall (Fig. 1A). Cells have two stellate chloroplasts with a central pyrenoid. Vegetative cells are 29-30 μm in width, 30-50 μm in length. Sexual reproduction is scalariform (Fig. 1B). Zygospores are formed in receptive (female) gametangia that remain cylindrical or slightly enlarged on the conjugating side (Fig. 1C). Zygospores are spherical or ellipsoid, 25-35 μm wide, and 30-50 μm long. Median spore wall is smooth
and yellow-brown at maturity.
Remarks. Tyndaridea insignis Hassall was transferred to the genus Zygnema by Kutzing (1849) based on the vegetative cell size and the shape of zygospores. Jao (1935) provided characteristics of additional features such as lateral or scalariform conjugation, shape of fertile cells, shape and size of zygospores, ornamentation and color of median spore wall. Transeau (1951) observed the occurrence of aplanospores.
Korean specimens correspond well with the previous descriptions of the species, except aplanospores and lateral conjugation. Korean Z. insigne has a vegetative cell width of 25-30 μm, zygospores which are formed in receptive gametangia, mostly cylindrical female gametangia, spherical or ellipsoid zygospores, and a smooth median spore wall. It is common in the fresh water systems in Korea.
Z. insigne has a similar vegetative cell width to Z. stellinum (Vaucher) Agardh and Z. vaginatum (Vaucher) Agardh (Randhawa 1959, Kadlubowska 1984), as well as similar scalariform conjugation and zygospores formed in receptive gametangia, and color of mesospores. However, Z. stellinum has enlarged female gametangia, ovoid zygospores and a scrobiculate median spore wall, and Z. vaginatum contains slightly enlarged female gametangia, globose to ovoid zygospores and a verrucose-tuberculate median spore wall. This species was previously described
in Korea (Chung 1968, 1993), and has also been reported in Japan (Yamagishi 1965) and China (Jao 1935).
De Bary 1858, p. 77, Pl. I, Figs 7-14; Czurda 1932, p. 119, Fig. 123; Transeau 1951, p. 32; Randhawa 1959, p. 234, Fig. 175; Kadlubowska 1984, p. 174, Fig. 240.
Specimens examined. Stream, Yeongdeok, Korea (Oct 29, 2004); Ditch water, Taean, Korea (Oct 25, 2007); Gungnam pond, Buyeo, Korea (Apr 30, 2008); Rice field, Eumseong, Korea (May 9, 2008); Rice field, Jecheon, Korea (May 15, 2008).
World distribution. British Isles, China, Europe, Greenland, Iceland, Japan, and the United States of America.
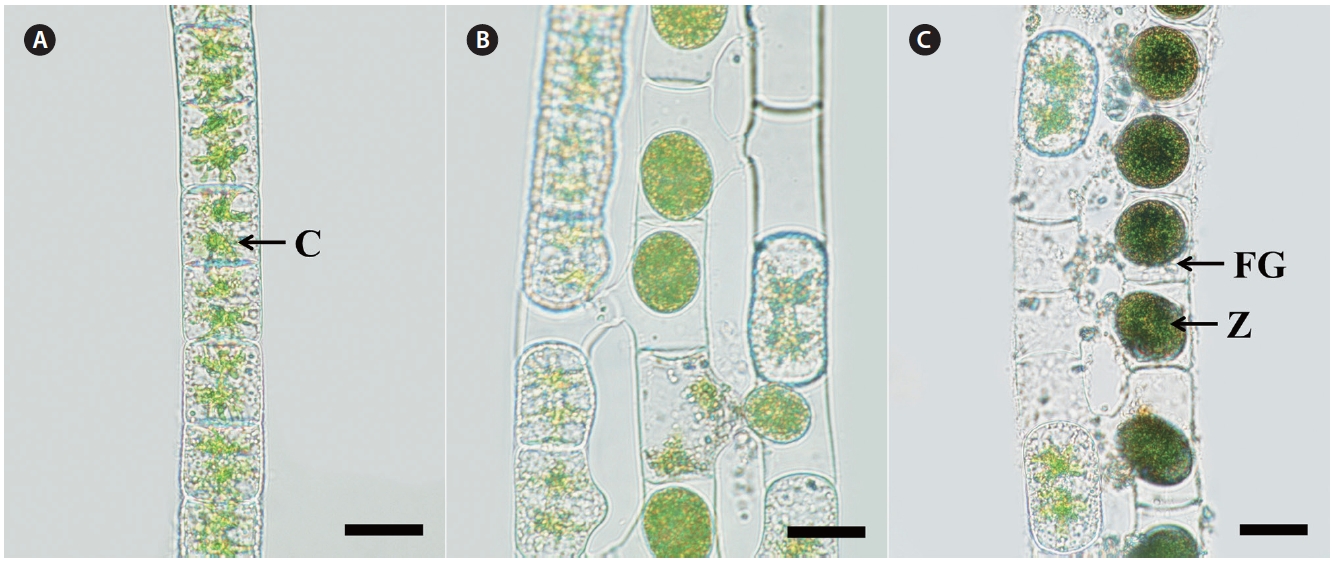
Morphology. Plants are unbranched filamentous of cylindrical cells with plane end wall (Fig. 2A). Cells have two star-shaped chloroplasts and each chloroplast has a central pyrenoid. Vegetative cells are 22-30 μm in width, and 27-100 μm in length. Sexual reproduction is scalariform (Fig. 2B). Zygospores are formed in only one of the gametangia that remain enlarged on the conjugating side (Fig. 2C). Zygospores are spherical, 24-43 μm wide and 24-43 μm long. The median spore wall is smooth and yellowbrown at maturity.
Remarks. Z. leiospermum was described by De Bary (1858) based on the vegetative cell size and features of zygospores. Czurda (1932) added features such as lateral conjugation, shape of fertile cells, shape and size of zygospores, ornamentation and color of median spore wall.
Korean specimens agree well with the description of Z. leiospermum except aplanospores. It is distinguished by the formation of zygospores in receptive gametangia, enlarged female gametangia, ovoid or spherical zygospores, and a smooth median spore wall. This species showed remarkable variations in the size of vegetative cells and zygospores in Korean populations. Genetic variation of Korean specimens, despite the conserved psbA gene in freshwater algae (Boo et al. 2010), is an interesting result that warrants further study.
Z. leiospermum is comparable to Z. hausmannii (De Notaris) Czurda and Z. luteosporum Czurda in terms of the width of the vegetative cells, scalariform conjugation, formation of zygospores in receptive gametangia, and yellow-brown mesospore (Transeau 1951, Kadlubowska 1984). Z. hausmannii has enlarged female gametangia, and scrobiculate mesospores with large (7-9 μm) diameter of pits, and Z. luteosporum contains cylindrical or slightly enlarged female gametangia, ovoid zygospores and a scrobiculate median spore wall with pits of less than 2 μm diameter. This species was previously listed in Korea without description or illustration (Yi 1980, Kim and Chung 1982), and has also been reported in Japan (Yamagishi 1965).
The 923 nucleotides of the psbA gene were determined for 22 isolates of Zygnema and its relatives. Of these, 34 positions (3.7%) were variable, and 178 positions (19.3%) were parsimoniously informative. Nucleotide substitutions for Zygnema consisted of 2.22 times more transitions than transversions.
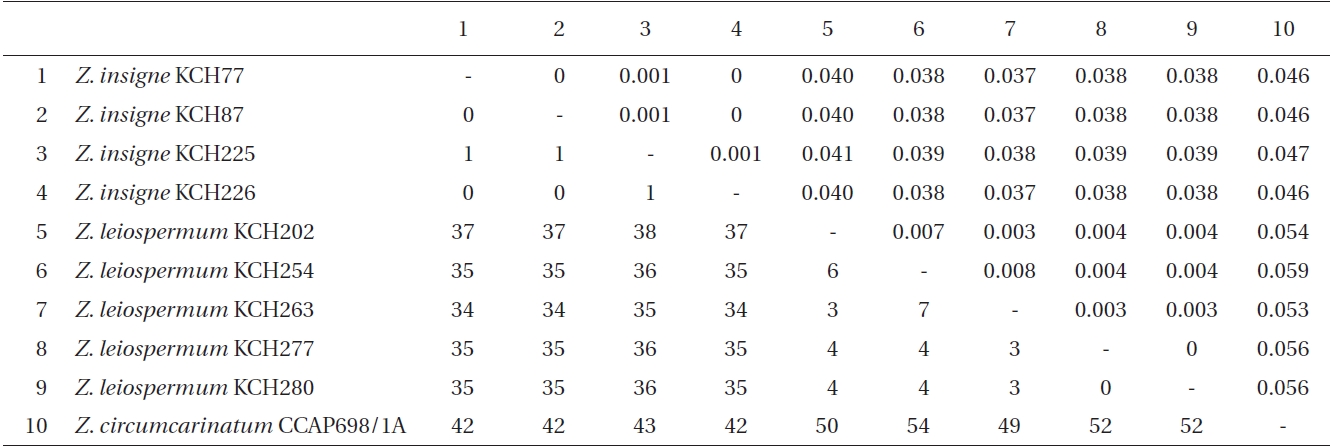
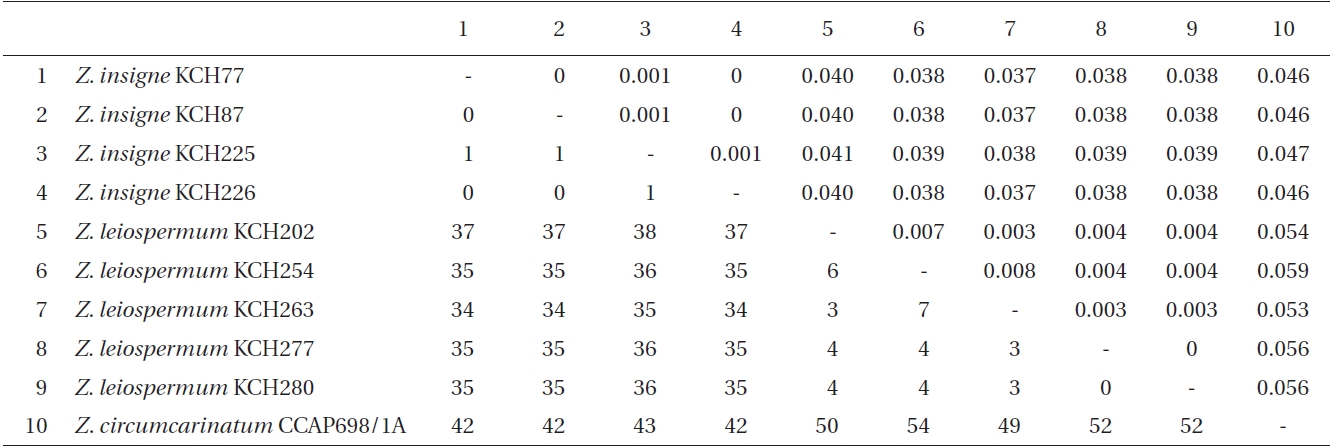
Within ingroup (Table 3), intraspecific pairwise divergence ranged from 0 to 0.1% within Z. insigne and to 0.8% within Z. leiospermum. However, two isolates of Z. circumcarinatum, one from SAG698-1a and the other from CCAP698/1A, differed by 43 bp (4.7% sequence divergence, not shown here). Interspecific pairwise divergence ranged from 3.7 to 4.1% (34-38 bp) between Z. insigne and Z. leiospermum. The p-divergence between Z. circumcarinatum CCAP698/1A and Z. leiospermum was slightly greater (5.3-5.9%, 49-54 bp) than that between Z. circumcarinatum CCAP698/1A and Z. insigne (4.6-4.7%, 42-43 bp). Outside from Zygnema, intraspecific pairwise divergence was 0 within Spirogyra decimina, S. ellipsospora, and Mougeotia sp. 1. The sequence divergence for psbA gene within Spirogyra ranged from 1.3% (between S. decimina and Spirogyra sp. KCH275) to 3.9% (between S. ellipsospora and Spirogyra sp. KCH251), and within Mougeotia ranged from 2.8% (between Mougeotia sp. 2 and Mougeotia sp. 1) to 10.9% (between M. transeaui and Mougeotia sp. 1). Sequence divergence between Zygnema and outgroup ranged from 7.5% between Z. insigne and Spirogyra sp. KCH275 to 12.3% between Z. insigne and Mougeotia transeaui.
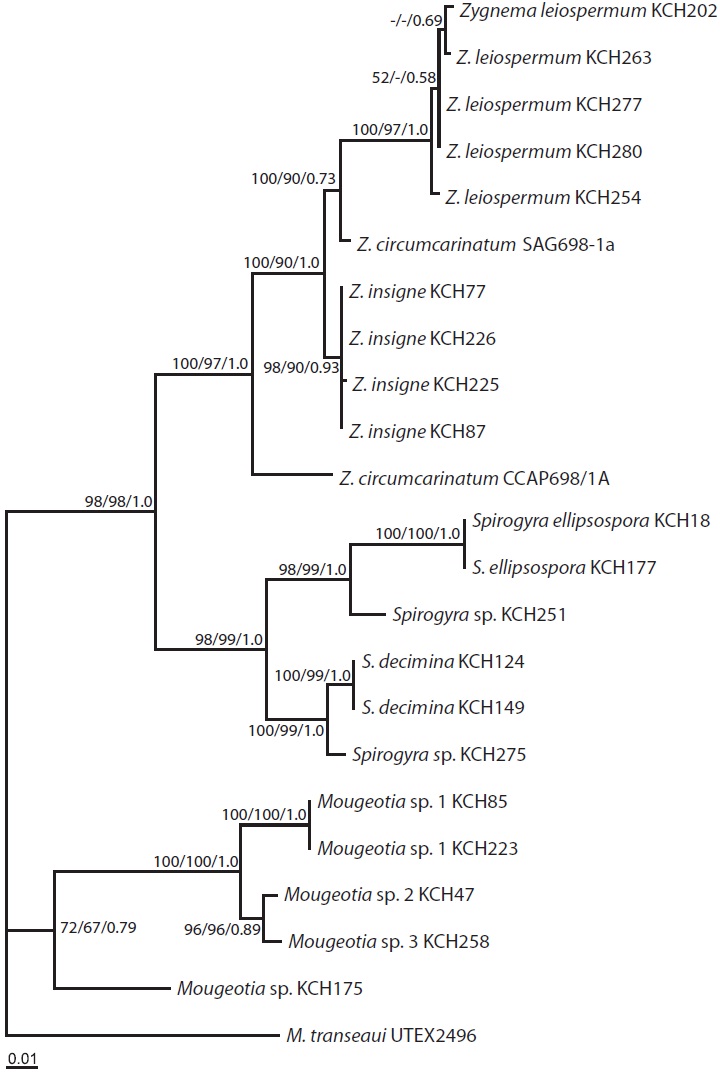
The ML tree (Fig. 3) was identical to the single most parsimonious tree (tree length = 1,182 steps, consistency index = 0.589, and retention index = 0.828). The genus Zygnema formed a well-resolved clade (100% for MP and 97% for ML, and 1.0 Bayesian posterior probability for BA), being subdivided into four lineages: Z. insigne, Z. leiospermum, and two lineages of Z. circumcarinatum. Z. insigne and Z. leiospermum, both from Korea, were clearly distinct within the clade.
This is the first psbA report on the phylogenetic relationships of Zygnema in the family Zygnemataceae. We recognized only two species from Korea, Z. insigne and Z. leiospermum, despite the collection of samples from many water systems over more than three years (2007- 2010). However, 11 species of Zygnema were reported in previous floristic studies in Korea (Chung 1970, Yi 1980, Wui and Kim 1990). The species not collected through the present study are Z. carinthiacum, Z. decussatum, Z. pectinatum, Z. peliosporum, Z. shigaense, Z. stellinum, Z. sterile, Z. vaucherii, and Z. cruciatum. Most of these species were described in morphology, but they were only reported for one or two locations. The stark discrepancy between our results, with extensive sampling and only a limited species collection, and the previous reports, may be due, at least in part, to misidentification of field-collected materials in previous floristic studies, or due to a reduction in the survival and reproduction of Zygnema species in Korean water bodies.
The topologies of the psbA trees are basically congruent in MP, ML, and BA. The monophyly of Zygnema was strongly supported (100% for MP, 97% for ML, and 1.0 for BA), as was in previous rbcL (McCourt et al. 1995, 2000), SSU rDNA (Gontcharov et al. 2003), both rbcL + SSU datasets (Gontcharov et al. 2004, Hall et al. 2008), and rbcL + cox3 datasets (Stancheva et al. 2012).
In our psbA sequence analyses, taxa of Zygnema consisted of four groups, in which two Korean species were included: Z. insigne and Z. leiospermum. The intraspecific divergence of Z. insigne was up to 0.1%. However, Z. leiospermum was variable in filament width and length, and in psbA sequences (up to 0.8%). The high pairwise divergence may be reflected either from morphological variation within the Zygnema species (Stancheva et al. 2012) and / or different ecological niches (Randhawa 1959).
Z. insigne and Z. leiospermum shared apomorphic characters such as scalariform conjugation, zygospores formed in receptive gametangia, and yellow-brown and smooth mesospores at maturity. However, they differed chiefly in their vegetative cell size, shape of female gametangium, and shape and dimensions of zygospore. Z. insigne is characterized by having cylindrical or slightly enlarged female gametangia, spherical to ellipsoid zygospores, and a broad vegetative cell width of 29-30 μm. Z. leiospermum is characterized by having enlarged female gametangia, spherical zygospores, and a narrow filament of 22-24 μm (Kadlubowska 1984).
Z. circumcarinatum CCAP698/1A and the two Korean species were similar in terms of their scalariform conjugation, spherical zygospores, and yellow-brown mesospores at maturity. Despite their similarity to the two species from Korea, Z. circumcarinatum is characterized by having 20-22 μm vegetative cells, zygospores formed in the conjugating tube, spherical or compressed-spherical zygospores, pitted and yellow-brown mesospore at maturity (Transeau 1951, Kadlubowska 1984).
Z. circumcarinatum SAG698-1a (Turmel et al. 2005) and CCAP698/1A analyzed in the present study markedly differed by 43 bp (4.7% p-distance) and hence, are not grouped together (Fig. 3). Based on the comparisons of rbcL sequences among four strains of Z. circumcarinatum from the Microalgae and Zygnemophyceae Collection of Hamburg (MZCH), UTEX, and SAG698-1a, Stancheva et al. (2012) suggested that the strain SAG698-1a (Turmel et al. 2005) is not Z. circumcarinatum, but is another species of the genus.
A single monophyletic Spirogyra clade was sister to Zygnema clade (98% for MP and ML and 1.0 for BA), as was present in previous rbcL (McCourt et al. 1995, 2000), and both rbcL + SSU datasets (Hall et al. 2008). In our psbA analyses, taxa of Spirogyra were divided into two major clades (98% for MP, 99% for ML, and 1.0 for BA), one containing S. ellipsospora and Spirogyra sp. KCH251, and the other consisting of S. decimina and Spirogyra sp. KCH275. A previous study on the phylogeny of 15 Spirogyra species from Korea, based on plastid rbcL gene, revealed four major clades within the genus (Kim et al. 2006). The four clades are morphologically supported, although the inter-cladal relationships have not been resolved (Kim et al. 2006).
In addition, four Mougeotia species were confirmed to occur in Korea. The results reveal the molecular taxonomy of Korean Mougeotia in which six morphological species of the genus have been reported (Chung 1993). Only one species of the genus was investigated for rbcL in Korea (Kim et al. 2006). Taxonomic revision of Spirogyra and Mougeotia from Korea are the next aim of this study group.
In conclusion, we confirmed two species of Zygnema, Z. insigne and Z. leiospermum from Korea, based on combined study of morphology and psbA gene sequences. Although vegetative filaments were variable in Korean Zygnema species, morphological features such as the size of vegetative cells, details of sexual reproduction, shape and dimensions of spore, and ornamentation and color of spore proved to be useful characteristics for identifying Zygnema species. Further taxon sampling may confirm more Zygnema species in Korea, and analyses of the fast evolving genes such as tufA may be needed for a better understanding of the genus.