In 2009, a systemic megalocytivirus infection associated with high mortality was detected for the first time in cultured starry flounder Platichthys stellatus in Korea. Diseased starry flounder had pale bodies and gill coloring and enlarged spleens. Histopathological examinations revealed basophilic enlarged cells in various organs of diseased starry flounder. Polymerase chain reaction (PCR) was performed on tissue samples using three published primer sets developed for the red sea bream iridovirus. PCR products were detected for all primer sets, except 1-F/1-R, which are registered by the World Organization for Animal Health (OIE). The part of the gene corresponding to the full open reading frame encoding the viral major capsid protein (MCP) was amplified by PCR. PCR products of approximately 1,581 bp were cloned, and the nucleotide sequences were analyzed phylogenetically. The MCP gene of the starry flounder iridovirus, designated SFIV0909, was identical to that of the turbot reddish body iridovirus (AB166788).
The starry flounder,
Members of the family Iridoviridae are an emerging group of viral pathogens that are threatening the aquaculture industry; they have caused large economic losses globally. Iridoviruses are large (approximately 120-350 nm) double-stranded DNA viruses that can infect invertebrates and poikilothermic vertebrates, including insects, fish, amphibians, and reptiles (Williams et al., 2005). The family Iridoviridae comprises five genera, including
In 2009, a disease outbreak occurred in starry flounder farms in Gyeongsangnam-do, Korea, and caused mortality rates of up to 60%. During biopsies of diseased fish, enlarged cells were observed in stamp samples from enlarged spleens, which is suggestive of RSIV disease. In addition, we attempted to detect portions of the iridovirus genome in the spleens and kidneys of diseased fish using PCR, as recommended by the World Organization for Animal Health (OIE) (World Organization of Animal Health, 2009); positive results were obtained only using the primer set 4-F/4-R, and not 1-F/1-R. This is the first study of iridovirus infection of farmed starry flounder and included taxonomical and histopathological analyses.
Diseased and healthy starry flounder were collected in September 2009 from a farm located in Gyeongsangnam-do, Korea. Mean fish weight was 22 g. The fish were transported with aeration to a laboratory at the Aquatic Animal Disease Control Division, National Fisheries Research and Development Institute, Busan, Korea, and were alive before examination. Affected fish were lethargic and had reduced appetite, pale body color, enlarged abdomen, and protruding eyes. Key internal characteristics of diseased fish included extremely enlarged spleen and kidney, and pale gills and/or liver.
After sampling some diseased starry flounder, the remaining fish were sacrificed and facilities were disinfected with sodium hypochlorite.
>
Viral DNA extraction and polymerase chain reaction
Total DNA was extracted from approximately 50 mg of spleen and kidney tissue from diseased starry flounder using a high-purity PCR template preparation kit (Roche, Indianapolis, IN, USA). PCR product quality and concentrations were determined by agarose gel electrophoresis and using a spectrophotometer (NanoVue; GE Healthcare, Little Chalfont, UK).
[Table 1.] Primers used in the present study
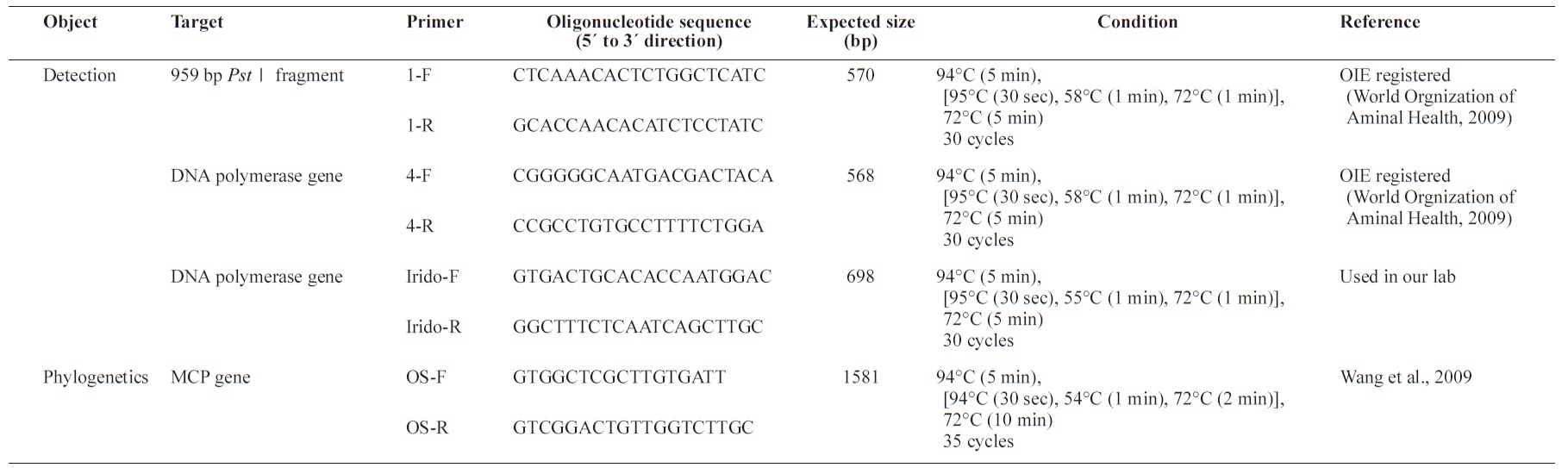
Primers used in the present study
For diagnosis of RSIV, three primer sets were synthesized for use in PCR assays. Two primer sets, 1F/1R and 4F/4R, were designed by Kurita and were registered in the list of the OIE (World Organization of Animal Health, 2009). The other primer set, IridoF/IridoR, was routinely used in our laboratory to detect RSIV. For the phylogenetic analysis, the OSF/OSR primer set was used (Table 1). Extracted genomic DNA (100 ng) was PCR amplified using Hot Start Ex
>
Sequencing and phylogenetic analysis of the major capsid protein (MCP) gene
The PCR products were purified using a gel purification kit (GeneAll, Seoul, Korea). The purified products were cloned with a TOPO-TA cloning kit (Invitrogen, Carlsbad, CA, USA) and subjected to nucleotide sequence analysis using an ABI PRISM 3730 XL DNA Analyzer with a BigDye terminator kit (Applied Biosystems, Inc., Foster City, CA, USA). Nucleotide sequences obtained from individual sequencing reactions were assembled using the Data Collection and Sequence Analysis software (Applied Biosystems, Inc.).
Sequences acquired in this study were compared with those from other megalocytiviruses that are available in the Gen- Bank/EMBL nucleotide database; alignment was performed using the software Genetyx v. 7 (Genetyx, Tokyo, Japan). Phylogenetic trees were constructed by neighbor-joining analysis with the software MEGA v. 4.0 (Department of Biology, Arizona State University, Tempe, AZ, USA).
>
Histopathological examinations
For histological studies, the spleen, kidney, gill, intestine, and stomach were removed from moribund and surviving starry flounder and immediately fixed in 10% neutral-buffered formalin. After fixation, standard histological procedures were used for tissue dehydration and paraffin embedding. Tissue sections were stained with hematoxylin and eosin (H&E). Stained sections were microscopically examined at magnifications ranging from 40× to 1000× (BX50F4; Olympus, Tokyo, Japan).
>
Isolation and identification of iridovirus from starry flounder (SFIV0909)
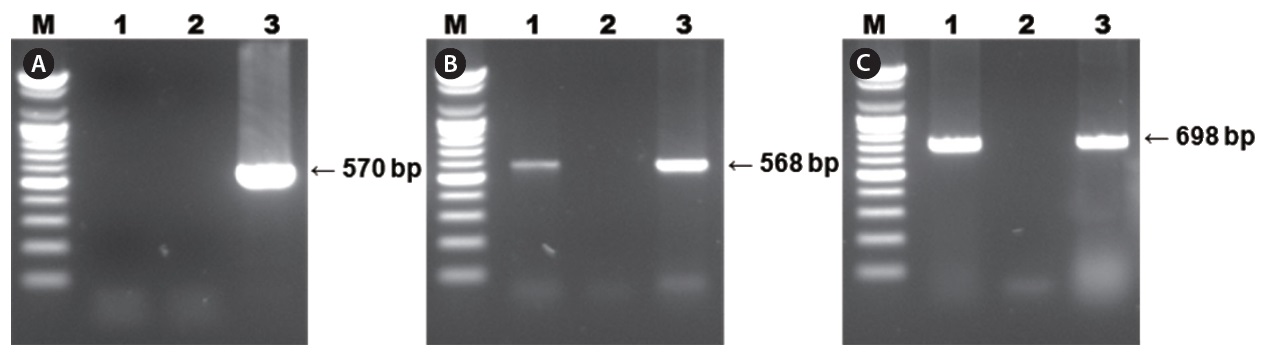
Diseased fish collected from the 2009 disease outbreak in Gyeongsangnam-do, Korea, were lethargic and showed anemia, petechiae of the gills, and enlargement of the spleen. The results of PCR using primer sets for diagnosing RSIV are shown in Fig. 1. Two specific PCR products were amplified from the spleen and kidney using the 4F/4R and IridoF/IridoR primers . However, the 1F/1R primer set, which was registered by OIE as RSIV-specific, did not amplify a product. The distilled water negative control showed no amplification. Parasites were absent in smears and wet mounts of infected fish, and bacteria were not consistently isolated from diseased fish.
Results of RSIV PCR using three primer sets are shown in Fig. 1. Specific PCR products corresponding to the 568-bp fragment were amplified using the 4F/4R primer set and products corresponding to the 698-bp fragment were amplified using the Irido-F/Irido-R primer set from all diseased starry flounders. However, PCR using the 1F/1R primer set, and the negative control, did not result in amplification of a product.
>
Analysis of the MCP nucleotide sequence
To determine the genetic characteristics of the iridovirus from starry flounder (SFIV0909), a full-length open reading frame (ORF) of the MCP of SFIV0909 was amplified by PCR. The ORF of MCP was 1,362 bp in length and encoded a protein that contained 453 amino acids. The nucleotide sequence
of SFIV0909 MCP was highly homologous to that of RSIV from other flatfish, such as turbot (urbot iridovirus [TBIV] AB166788 and TRBIV AY590687) and olive flounder (FLIVPH AY633992, FLIV-JSY AY633989, FLIV-M AY63398,
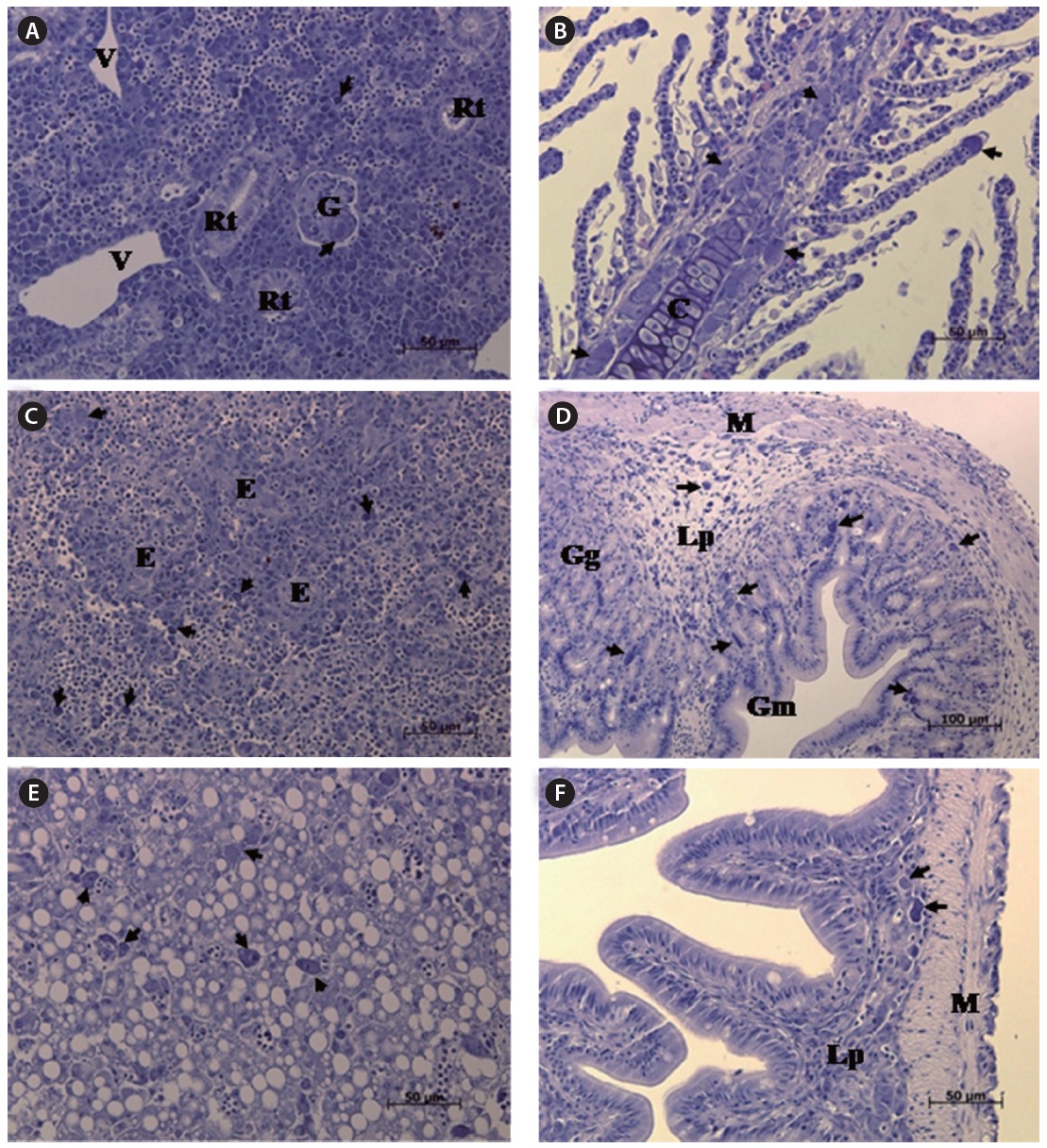
The most striking histopathological change in moribund starry flounder was the formation of basophilic enlarged cells in various tissues, including the spleen, kidney, gills, intestine, and stomach. In particular, the spleen displayed large numbers of enlarged cells within splenic pulps and sheathed tissue (Fig. 3). Severe, diffuse necrosis, accompanied by hemorrhage, was also evident in the spleen.
This report describes the identification and histopathological characterization of an iridovirus from diseased starry flounder. In Korea, megalocytiviruses were first reported as fish pathogens in 1998, when they caused mass mortality in rock bream,
Although iridovirus infections have been documented in many countries and various fish species, iridoviruses associated with starry flounder have not to date been reported. This is probably because the history of starry flounder culture is short; cultivation of starry flounder has begun to expand only recently. In Korea, starry flounder culture started in 2005 with the production of 50,000 seed fish. The annual production of farmed starry flounder began at 17 metric tons in 2007 and increased to 158 metric tons in 2008, 341 metric tons in 2009, and 466 metric tons in 2010 (Statistics Korea, 2011). Starry flounder aquaculture is thought to have continued to develop rapidly in subsequent years because the species tolerates a wide range of salinities, endures crowded environments and exhibits rapid growth (Ding et al., 2010). Nevertheless, information about the diseases of starry flounder remains scarce. The only previous study of a disease outbreak in starry flounder reported an infection caused by
The outbreak in 2009 described in this study was the first to be documented in cultured starry flounder in Korea. In our study, we found that while no apparent bacterial infection was observed in moribund fish, SFIV0909 was detected in all diseased fish examined, which suggests that SFIV0909 was the etiological agent of the outbreak. Some unexpected results were also found in the PCR screening for RSIV because the specific primer set 1-F/1-R, which is recommended by OIE, did not amplify SFIV0909 from these starry flounder. This primer set is the most frequently used for the detection of RSIV; however, other researchers also failed to detect a fragment using the same forward primer1-F (Shi et al., 2010). They found that PCR of the spleens and kidneys of turbot with red body syndrome yielded no specific product, and reported detection by only transmission electron microscopy (TEM). Therefore, the results from any single primer set should be interpreted with caution.
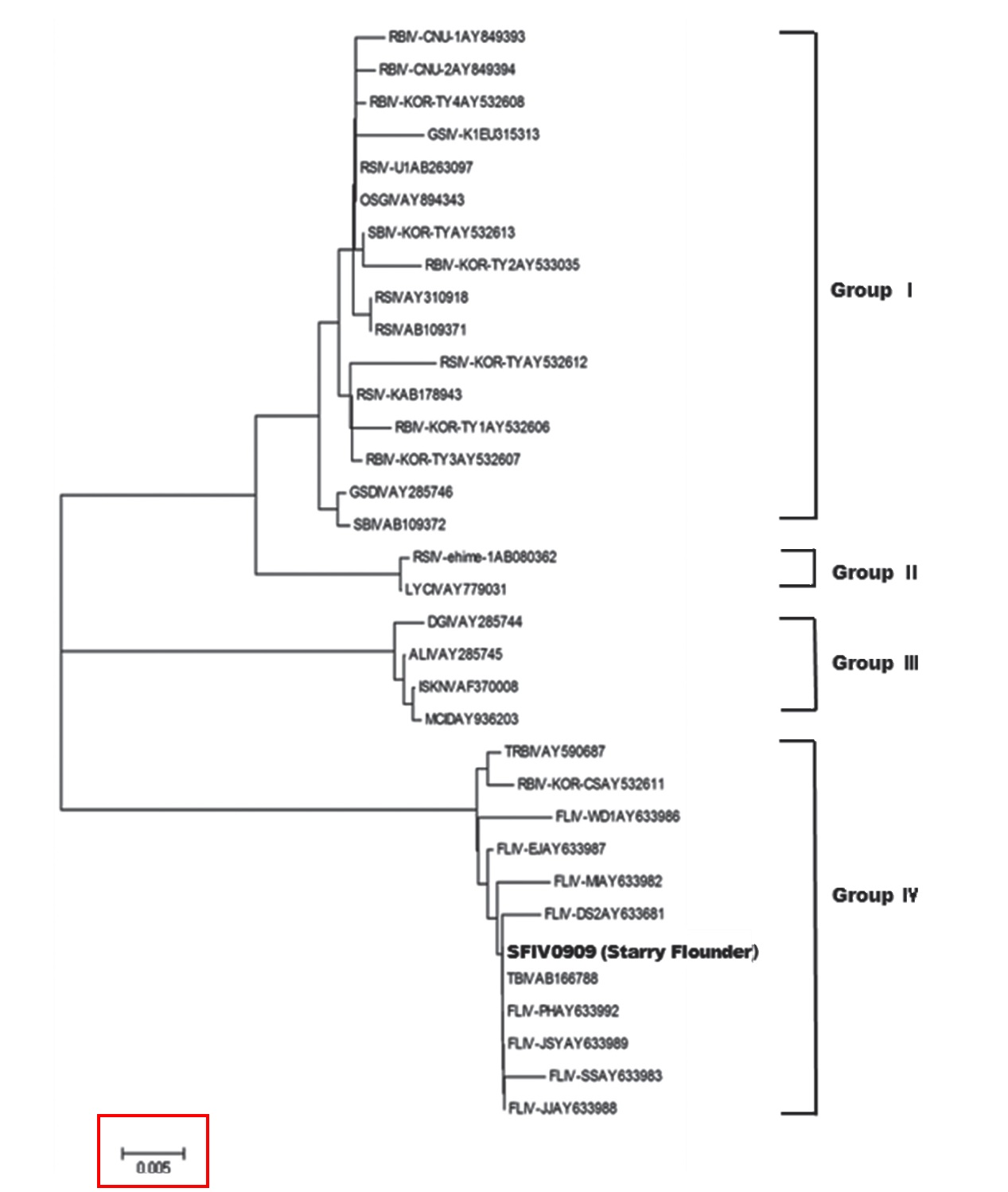
Megalocytivirus isolates have been divided into four genotypes based on their MCP gene sequences (Dong et al., 2010). Megalocytiviruses in groups I and II are derived mainly from perciform fishes, whereas all megalocytiviruses from freshwater fishes fall into group III. Viruses in group IV are derived mainly from Pleuronectiformes in Korea and China. To date, olive flounder
In this study, SFIV0909 was found to belong to megalocytivirus genotype IV, along with most of the iridoviruses that infect turbot and olive flounder. This study also found 100% homology with TBIV (AB166788) from turbot in Korea (Oh et al., 2006), suggesting that the starry FLIV is identical to the agent found in turbot in Korea, and that it evokes the same pathogenicity in flatfishes. Histopathological changes in starry flounder were similar to those in the turbot,
In conclusion, we detected for the first time a megalocytivirus, SFIV0909, in cultured starry flounder in Korea. Further work involving experimental inoculations of starry flounder with this virus should be completed to determine whether this iridovirus is the causative pathogen of the 2009 starry flounder outbreak. We found multiple indications that this virus was likely the causative agent, including the absence of other microbes and parasites in infected tissues and severe histopathological and cytopathological effects on various tissues and organs that were apparently induced by the virus. These results suggest that SFIV0909 causes serious systemic infection in starry flounder.