The presence of ostreid herpesvirus 1 (OsHV-1) and the percentage of viral DNA detected in Pacific oyster Crassostrea gigas adults were investigated monthly between May and November 2012 at three locations along the southern coast of Korea. Among 210 oysters examined by polymerase chain reaction (PCR) analysis, OsHV-1 DNA was detected in only one oyster collected in August. The low detection rate of OsHV-1 DNA was consistent with the lack of reported OsHV-1-associated disease in C. gigas cultured in Korea. The sequence of the present PCR product amplified with the C2/C6 primer pair was identical to that of OsHV-1 μVar except for one nucleotide, and the sequence amplified with Del36-37F2/Del36-37R showed a 605-bp deletion as in OsHV-1 μVar. Although these sequence data are insufficient to determine genotype, the results suggest that the herpesvirus detected was similar to OsHV-1 μVar. This is the first report on the presence of OsHV-1 in adult Pacific oysters cultured in Korea.
Since Farley et al. (1972) first described a herpes-type virus in the Eastern oyster
Korea is among the top three countries in the world for production of Pacific oysters. However, as oyster farms are concentrated on the southern coastal area of Korea, an outbreak of highly infectious disease at a farm can bring highly disastrous results in the Korean oyster aquaculture industry. Although Moss et al. (2007) reported the presence of molluscan herpesvirus from
The excised gill and mantle were lysed by adding 500 μL of lysis buffer containing Proteinase K and incubated in a shaking water bath at 60℃ for 3 h to extract total DNA. Then, 250 μL of 6 M NaCl was added to each tube, followed by vigorous shaking of the tubes 20 times and a 10-min incubation on ice. The supernatants were recovered by centrifugation at 10,000
The primers used are listed in Table 1. Primary PCR reactions were conducted with primer pairs of C13/C5 (open reading frame [ORF]4), C13/C6 (ORF4), and Del-F/Del-R (ORF35, 36, 37, and 38), and amplification conditions were one cycle of 3 min at 95℃ (initial denaturation), 30 cycles of 30 s at 95℃, 30 s at 55℃, and 30 s at 72℃, followed by 7 min at 72℃. Primer pairs used for nested PCR were C2/C4, C2/C6, and Del36-37F/Del36-37R, respectively, and 0.2 μl of each primary PCR product was used as the template. The PCR conditions for the nested PCR were the same as those for the
[Table 1.] Oligonucleotides used in this study

Oligonucleotides used in this study
primary PCR. PCR products were analyzed on a 1% agarose gel containing Midori Green Advanced DNA stain (Nippon Genetics, Duren, Germany) for visualization, purified using a gel purification kit (Nucleogen, Seoul, Korea), subcloned into pGEM-T Easy Vector (Promega, Madison, WI, USA), and sequenced. Multiple sequence alignments were generated using the CLUSTALW 1.8 program.
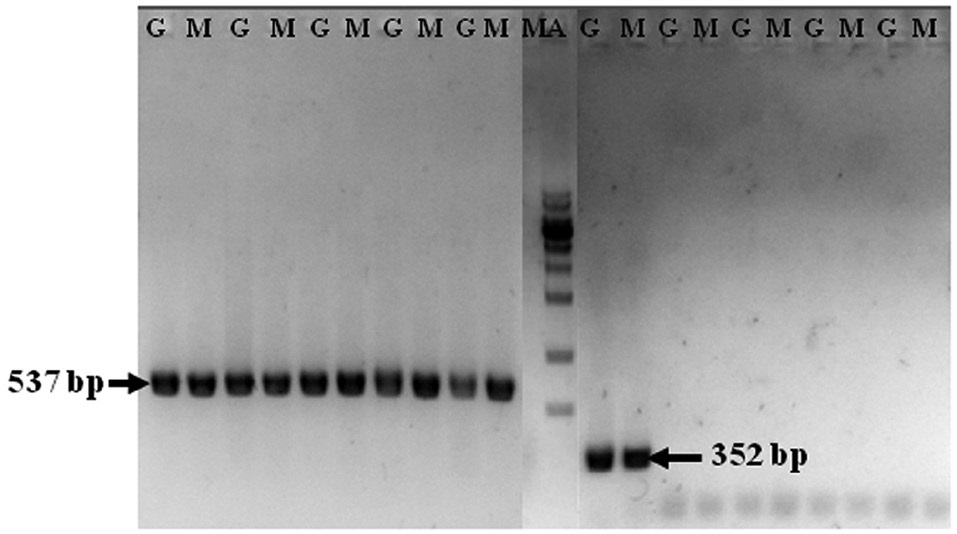
In total, 210 Pacific oysters were PCR-analyzed to detect OsHV-1 DNA. The band corresponding to the partial 18S ribosomal RNA gene was detected from all analyzed specimens (Fig. 1). No amplified band was identified in any of the samples by primary PCR with the primer pairs for amplifying OsHV-1 DNA. However, nested PCR with the C2/C4 primers amplified a band of the expected size (352 bp) from only one oyster (Fig. 1), which was collected from Tongyeong in August; thus, the percentage of OsHV-1 DNA detection in
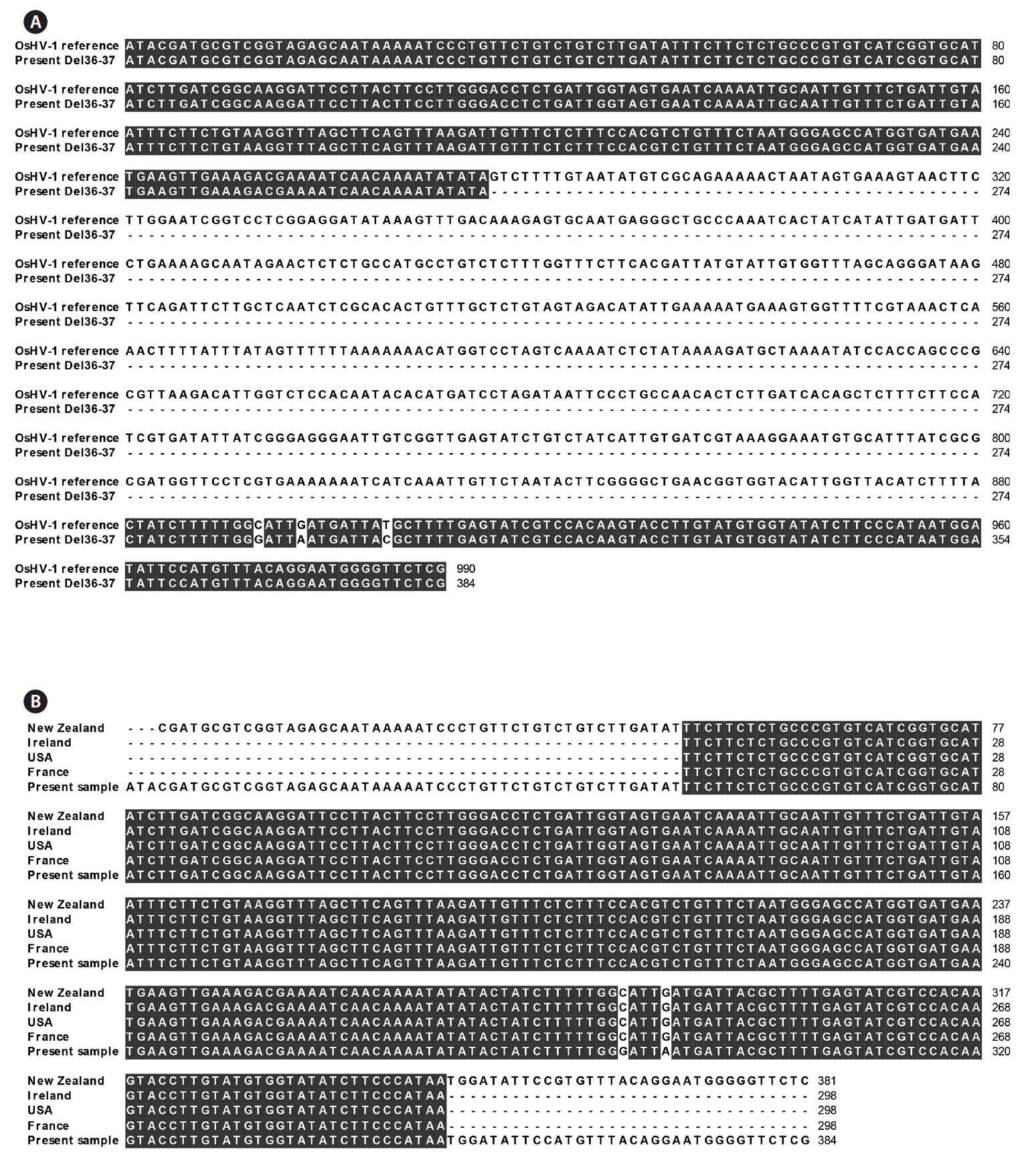
The sequence of the C2/C6 PCR product of the positive sample showed deletion of 13 consecutive nucleotides, and another 11 nucleotides were sporadically changed or deleted when compared to the OsHV-1 reference sequence (Fig. 2). Only one G (Fig. 2) was deleted in the present sample compared to that in the OsHV-1 μVar sequence.
Nested PCR with the Del36-37F/Del36-37R primers amplified a band of expected size (384 bp) that represented a deletion of the middle region of the gene corresponding to 605 nucleotides in the OsHV-1 reference sequence (Fig. 3A). A comparison of the present sequence with those recorded in other countries was identical except for two nucleotides (Fig. 3B).
We report for the first time the presence of OsHV-1 from Pacific oyster adults in Korea. Although OsHV-1 has been associated with mass mortalities of Pacific oyster larvae or spat, adult oysters asymptomatically carrying the virus could have the potential to transmit OsHV-1 to
According to the results of Moss et al. (2007),
One of the main differences in the nucleotide sequence of OsHV-1 μVar from that of OsHV-1 reference is a partial deletion of the microsatellite zone of the region amplified with the primer pair C2 and C6 (Segarra et al., 2010). The deleted nucleic acids included three consecutively repeated “CTA,” followed by a “CTGA.” The C2/C6 sequence of the present OsHV-1 showed the same deletion and was identical with that of OsHV-1 μVar except for one nucleotide, whereas the previously reported OsHV-1 from China and Japan showed four and three different nucleotides, respectively, compared to that of OsHV-1 μVar. Renault et al. (2012) reported that all specimens identified as OsHV-1 μVar show a large deletion (605 bp) of the amplicon with the primer pair Del36-37F2 and Del 36-37R, and the present virus specimen also had the same deletion. Considering the full genome size of OsHV-1, the present sequence data are insufficient to determine the genotype; however, our results suggest that the herpesvirus detected in this study was similar to OsHV-1 μVar.