The Javanese ricefish Oryzias javanicus is closely related to the Japanese ricefish O. latipes, a popular model organism. However, unlike O. latipes, O. javanicus is capable of hyperosmoregulation and can tolerate saltwater (Koyama et al., 2008). Due to this ability, previous studies have suggested the use of this euryhaline ricefish species as a potential indicator species for the ecotoxicology of brackish and marine environments (Yu et al., 2006; Woo et al., 2006, 2009). This model fish species has the same traits observed in most small, egg-laying Oryzias; year-round spawning, easy to culture in a laboratory, and a short-generation time (Koyama et al., 2008; Song et al., 2010). In addition to this, the Javanese ricefish is mostly transparent throughout its lifecycle, which is especially useful in transgene-based heterologous expression assays using fluorescent or colorimetric reporters. However, despite having all of these useful traits, the Javanese ricefish has not been widely used in transgenic investigations.
The cytoskeletal beta-actin (β-ACT) gene (actb), a member of the actin multigene family, encodes a non-muscle, cytoskeletal actin protein that is ubiquitously distributed in most animal cell types. The β-ACT is highly conserved in the animal kingdom and plays essential roles in maintaining cytoskeletal structure, cellular mobility, cell division, and contractile processes (Reece et al., 1992). In addition, actb is a housekeeping gene and due to its essential role, actb mRNA expression has often been used as a versatile invariant control for gene expression studies (Andreassen et al., 2005; Cao et al., 2007). More importantly, previous studies have demonstrated that the regulatory region of actb has a high capacity to drive efficient expression of downstream foreign gene(s) both in vivo and in vitro (Noh et al., 2003; Brooks et al., 2007; Yu et al., 2010). New functional fish strains with novel phenotypes, such as fast-growth (Nam et al., 2008) and enhanced immunity (Ruiz et al., 2008; Lin et al., 2010), have been generated using actb promoters. Furthermore, the ability to label the whole organism during its entire lifecycle using ubiquitous promoter-driven fluorescent transgenes can provide a unique approach for studying cell lineage and migration, transgene silencing, and tissue regeneration (Hsiao and Tsai, 2003; Burket et al., 2008).
We isolated and characterized the gene and promoter for actb in O. javanicus, to develop a heterologous expression system for this euryhaline fish. To this end, we determined the structural features of mRNAs, the genomic gene, and the upstream regulatory region; examined tissue and developmental expression; and evaluated the ability of the actb promoter to drive heterologous expression of a fluorescent reporter in microinjected embryos.
The Javanese ricefish specimens used in this study were a laboratory strain maintained at the Institute of Marine Living Modified Organisms (IMLMO), Pukyong National University, South Korea. Fish were maintained in water at a concentration of 15 g salt/L water (15 ppt) and water temperature was kept at 25-27℃ throughout the experiments. Spawning conditions, egg collection, and embryonic development monitoring were as outlined in previous reports (Song et al. 2010; Cho et al., 2011). Genomic DNA was purified from the fin or whole fry following a conventional SDS/proteinase K method (Cho et al., 2011). Total RNA extraction was performed using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. Reverse transcription from purified total RNA was performed using the Omniscript Reverse Transcription Kit (Qiagen) according to the manufacturer’s manual.
We performed an expressed sequence tag (EST) analysis, using a Javanese ricefish whole-body cDNA library (constructed with a Lambda ZAP cDNA Synthesis Kit; Stratagene, La Jolla, CA, USA) to isolate clones showing significant homology with previously known vertebrate β-ACTs (unpublished data). Selected ESTs (9 out of a total 4,500 ESTs examined) were assembled into contigs using Sequencher 4.2 (Gene Codes, Ann Arbor, MI, USA), and a full-length, continuous sequence of the actb cDNA was validated by reverse transcription-PCR (RT-PCR) isolation from whole-body total RNA, using RT-PCR primers OJβ-ACTc FW (5´-GTCACACACAGCTTGTGCGGGATA-3´) and OJβ-ACTc FW RV (5´-CAAGTCGGAACACATGTGCACTT-3´). Amplified RT-PCR product was purified using a QIAEX II Gel Extraction Kit (Qiagen) and cloned into the pGEM T-easy vector (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Inserts of six randomly chosen recombinant clones were sequenced in both directions by primer walking to determine the representative cDNA sequence of each gene.
Based on the cDNA sequence, genomic fragments of actb were isolated by PCR using the OJβ-ACTc forward and reverse primers (as above), gel-purified, and TA cloned as described above. Eight randomly chosen RT-PCR clones for each gene were sequenced by primer walking to determine representative genomic sequences. To obtain the 5´-flanking region of each gene, genome walking was performed using the GenomeWalker Universal Kit (Clontech Laboratories Inc., Mountain View, CA, USA). The continuous fragment spanning from the 5´-flanking region to the 3´-untranslated region (3´-UTR) was re-isolated by PCR using OJβ-ACTp 1F (5´-AGCCTGATAGTGACGCTTCA-3´) and OJβ-ACT gR (5´-AAGTCGGAACACATGTGCAC-3´), and its sequence was confirmed.
Prediction of the open reading frame (ORF) and deduction of amino acid sequence was carried out using NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Multiple sequence alignments at the nucleotide and amino acid levels were performed using ClustalW (http://align.genome.jp) to examine homology with representative orthologues. The alignment was imported into GeneDoc (http://www.nrbsc.org/gfx/genedoc/) for optimization and to calculating sequence identity. Theoretical molecular weight (kDa) values and isoelectric point (pI) values using the deduced amino acid sequence were calculated using ExPASy ProtParam (http://web.expasy.org/protparam/). Organization of the genome was compared to previously known vertebrate orthologue genes. Potential transcription factor (TF) binding motifs in the 5´-flanking upstream region of O. javanicus actb were predicted using TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME) and TF searches (http://www.cbrc.jp/research/db/TFSEARCH.html).
For adult tissues and developing embryos, the distribution patterns as well as basal expression levels of actb transcripts were assessed by real-time RT-PCR. Ten tissue samples (from the brain, eye, fin, gill, heart, intestine, kidney, liver, skeletal muscle, and spleen) were taken from 12 healthy individuals (average body weight, 1.4 ± 0.3 g), frozen on dry ice, and stored at -85℃ until used. A total of 300 embryo samples were taken at 13 different developmental stages (just fertilized, eight-cells, morula, blastula, gastrula, neurula, foursomite stage, heartbeat stage, retinal pigmentation stage, gill blood vessel formation stage, visceral blood vessel formation stage, spleen development stage, and just-hatching) and then also stored at -85℃ until total RNA extraction. For the control sample, the 18S rRNA gene (GenBank accession no. KC020115) of O. javanicus was cloned by PCR isolation. An aliquot (2 μg) of total RNA for each sample was reverse transcribed into cDNA using an oligo-d(T)20 primer and an O. javanicus 18S rRNA reverse primer (OJ18S-RV) at final concentrations of 1.0 μM and 0.05 μM, respectively. The RT product from each sample was diluted 4-fold (for actb) and 40-fold (for 18S rRNA control) with sterile distilled water, and 2 μL diluted cDNA template was used in a real-time PCR. Realtime PCR was performed using the 2× iQ SYBR Supermix (Bio-Rad, Hercules, CA, USA) and the iCycler iQ Real-Time Detection System (Bio-Rad). The β-ACT segment (amplicon, 254 bp) was amplified using the following primers: qOJβ-ACT 1F (5´-CAACTCATTGGCATGGCTTC-3´) and qOJβ-ACT 1R (5´-GCCTTCACAGAGGCAAATAC-3´). The 18S rRNA control (amplicon, 253 bp) segment was amplified using the following primers: qOJ18S RNA 1F (5´-TCCAGCTCCAATAGCGTATC- 3´) and qOJ18S RNA 1R (5´-AGAACCGGAGTCCTATTCCA- 3´). Based on the standard curve of each gene, using four log-dilutions of positive cDNA samples, PCR efficiencies were higher than 91%. Expression levels of actb mRNAs across the tissues and embryonic samples were normalized against the levels of the 18S rRNA control according to Kubista et al. (2006). We performed triplicate assays per cDNA sample, and differences in actb mRNA levels across tissue and embryonic samples were assessed by ANOVA, and by Duncan’s multiple range test with P = 0.05, using SPSS software version 10.1.3 (SPSS Inc., Chicago, IL, USA).
To construct a fluorescent reporter vector driven by the O. javanicus actb promoter, a 4.3 kb, 5´-upstream fragment, including the non-translated exon 1 and intron 1, was isolated by PCR using the following primers: OJβ-ACTp 2F (5´- ATGTCGAGAGCCTGATAGTGACGCTTCA- 3´) and OJβ-ACTp 2R (5´-ATACCGGT GGCTAAACTGGAAAAGAACA-3´). The 5’-ends of these primers were designed to include SalI (recognition sequence, GTCGAC) and AgeI (recognition sequence, ACCGGT) restriction sites, respectively, to facilitate downstream cloning. Amplified product was TA-cloned, spliced from the T-easy vector (Promega) by digestion with the SalI and AgeI restriction enzymes (New England Biolabs, Ipswich, MA, USA), and unidirectionally ligated upstream of the translation initiation codon (ATG) of the red fluorescent protein (RFP) gene in the pre-digested pDsRed2-1 plasmid vector (Clontech Laboratories Inc.). The resultant RFP vector was called pOJβ-actRFP (8.4 kb). Transient, heterologous expression assays were performed with microinjection of pOJβ-actRFP into O. javanicus embryos. Circular pOJβ-actRFP was resuspended in an injection buffer (10 mM Tris-Cl, 0.1 mM EDTA, pH 8.0; supplemented with 0.01% phenol red; Sigma- Aldrich, St. Louis, MO, USA) at a concentration of 50 μg/ mL. Fertilized eggs were collected from egg-laying females and immediately placed in an incubator at 15 ℃ until microinjected. Microinjection was conducted on one-celled embryos using a Narishige MMN-330 micromanipulator (Narishige Scientific Instrument Lab, Tokyo, Japan). After injection, the embryos were transferred to a 25 ℃ incubator containing 5 μm filtered water of 15 ppt salinity, adjusted with synthetic sea salt (Kent Marine, Acworth, GA, USA). Transient expression of RFP was monitored in developing embryos using NIS-Elements Microscope Imaging Software and AZ100 Epifluorescence microscopes (Nikon Co., Tokyo, Japan).
O. javanicus actb cDNA is 2283 bp long as follows: an 81 bp 5´-UTR; 1,125 bp for a single ORF encoding a putative polypeptide of 375 amino acids (aa); 673 bp for the 3´-UTR, including stop codon (TAA); and a 29 bp poly(A)+ tail. A putative polyadenylation signal (AATAAA) is located 27 bp before the poly A+ tail (GenBank accession no. JQ905607). The predicted molecular weight and theoretical pI value of the β-ACT polypeptide are 41.7 kDa and 5.29, respectively. Overall, O. javanicus β-ACT shared common structural characteristics with vertebrate orthologues at both the nucleotide and amino acid levels. Multiple sequence alignments between the O. javanicus β-ACT polypeptide and teleostean orthologues show a high degree of secondary structure homology, sharing a high number of amino acid sequences with species belonging to the Oryzias genus (alignment not shown).
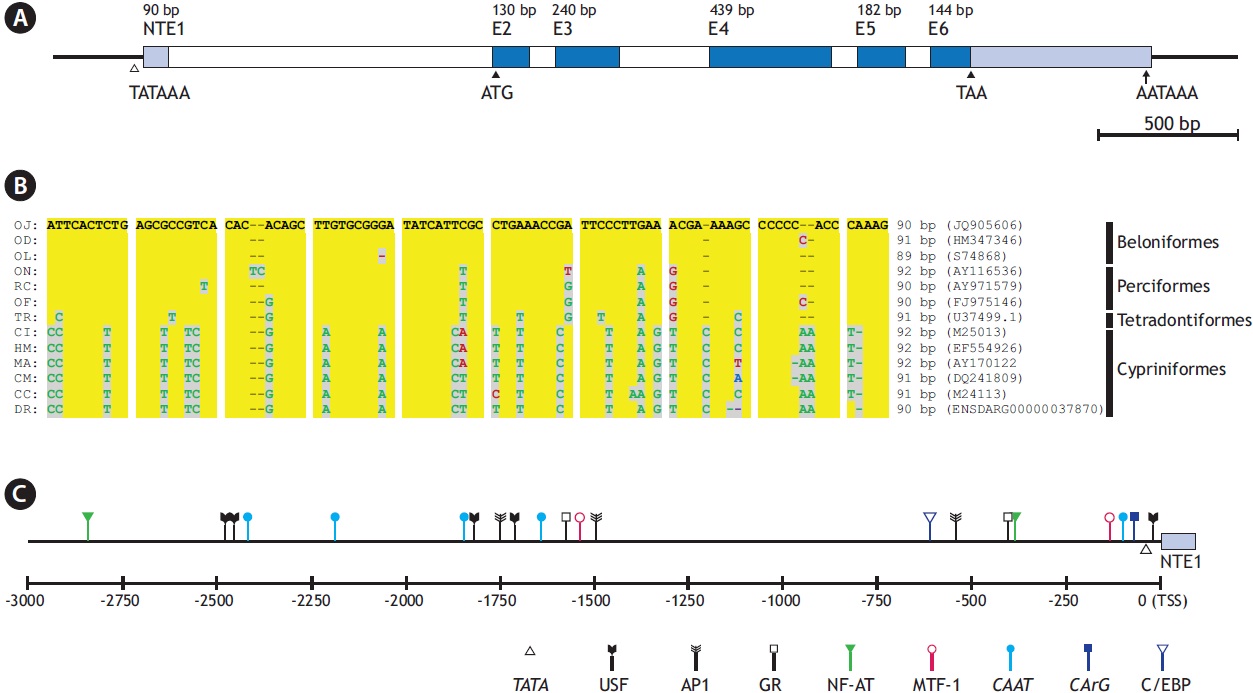
The O. javanicus actb gene consists of five translated exons (exons 2-6; 130, 240, 439, 182, and 144 bp, respectively), interrupted by four introns (introns 1-4; 92, 321, 74, and 81 bp, respectively) (GenBank accession no. JQ905606). As in other major actin genes in vertebrates, it also possesses a nontranslated exon (91 bp) beginning at -1,241 bp upstream from the ATG initiation codon, 8 bp after the 5´end of exon 2 (Fig. 1A). The intron, which follows the non-translated exon 1, is 1,144 bp long. The GT-AG exon/intron splicing rule is well conserved in all the boundary regions. Alignment of the putative O. javanicus non-translated exon (NTE1, 90 bp) with those from 12 representative teleosts showed that the fish actb genes shared considerable homology in their NTE1 sequences. The lengths of NTE1 in the other teleosts, calculated from the teleostean actb genes, ranged from 89 to 92 bp, and the overall similarity agreed with the known taxonomic appraisal (Fig. 1B). It is consistent with the known evolutionary history of the teleostean actin multigene family, consisting of gene
[Fig. 1.] Gene structure of Oryzias javanicus β-actin gene (actb). (A) Exon-intron organization including non-translated exon 1 (NTE1). (B) Multiple sequence alignment of NTE1 regions from representative teleosts. OJ, O. javanicus; OD, O. dancena; OL, O. latipes; ON, Oreochromis niloticus; RC, Rachycentron canadum; OF, Oplegnathus fasciatus; TR, Takifugu rubripes; CI, Ctenopharyngodon idella; HM, Hemibarbus mylodon; MA, Megalobrama amblycephala; CM, Cirrhinus molitorella; CC, Cyprinus carpio; DR, Danio rerio. (C) A schematic drawing to show the relative positions of transcription factor binding sites in -3 kb upstream regulatory region from transcription start site (TSS).
duplication and divergence from a common ancestral gene (Miwa et al. 1991; Kim et al. 2008).
The conserved characteristics are not restricted to the exon or coding regions, as O. javanicus actb also had common features in its regulatory region, including intron 1 following the NTE1. Bioinformatic analysis of the 5´ flanking region (i.e., the 3 kb upstream sequence from the putative transcription start site; TSS) and the 1.1 kb intron 1 predicts the presence of various TF binding sites including TATA and CAAT boxes (Fig. 1C). TATA (consensus sequence, TATAAA) and CAAT (CCAAT) signals are located at -29 bp and -92 bp from the putative TSS, respectively. A further three CAAT boxes were predicted in distal promoter regions, at -1,635 bp, -2,180 bp, and -2,415 bp from the TSS. Two CArG boxes (CC[W=A/ T]6GG) were identified: one in the proximal promoter region (CCTTTTATGG; -62 bp from the TSS) one in intron 1 (CCTTATATGG; -183 bp upstream from the ATG initiation codon) (Liu et al. 1990; Kim et al. 2008; Lee et al. 2009). Intron 1 of the vertebrate actb can function as an enhancer, and previous studies on fish actb regulators have shown its β-ACT regulatory role when it forms part of the transgene construct (Liu et al. 1991; Noh et al. 2003). A canonical E-box (CACGTG) is predicted 4 bp after the TATA box, and five non-canonical E-boxes (CANNTG) are identifiable in the distal promoter region, in a non-canonical form. Alongside these essential motifs, several TF binding sites related to the stress response are also expected. They include the following: sites targeted by nuclear factor for activated T-cells (NF-AT; consensus sequence, WGGAAAA); activator protein 1 (AP-1; TGA[S=G/C]T[M=A/C] A); CCAAT-enhancer binding protein (C/EBP; TT[D=A/G/T] NGNAA); glucocorticoid receptor (GR; half site, AGAACA or TGTTCT); and metal regulatory transcription factor 1 (MTF-1; TGC[R=A/G]CNC). Previous studies on fish β-ACT promoters have also reported similar TF profiles containing response factor binding sites (Kosuke et al., 2009; Cho et al., 2011), although functional characterization of the motifs have not been performed to date. Much of the literature claims that the fish actb gene might be modulated by stimulation and/ or the physiological states of the fish (Filby and Tyler, 2007; Small et al. 2008), and this may indirectly support our findings in the O. javanicus actb promoter.
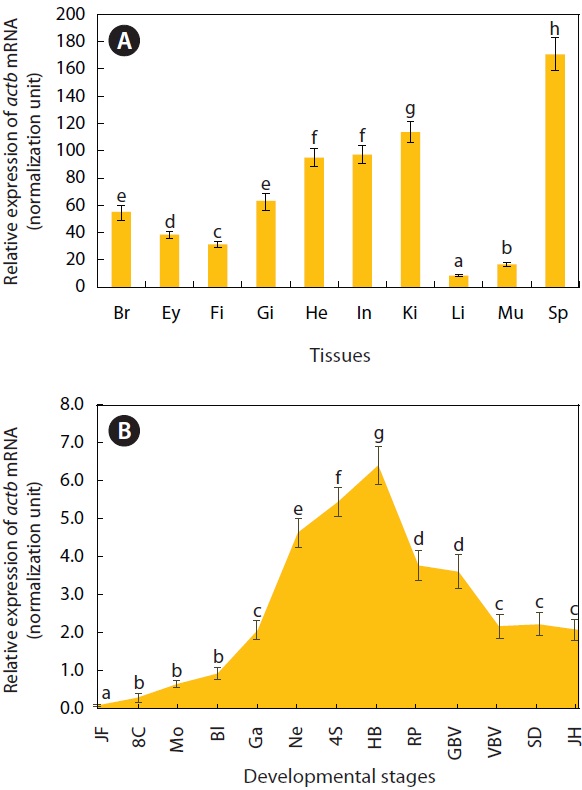
As expected, actb transcripts were ubiquitously detected in all tissues examined, based on the real-time RT-PCR assay (Fig. 2A). However, the basal expression levels were variable among tissue types when normalized against the 18S rRNA level for each tissue. The spleen showed the highest actb transcript level, with the liver and skeletal muscle exhibiting the lowest levels (P < 0.05). The kidney, heart, and intestine had
[Fig. 2.] Quantitative reverse transcription (qRT-PCR) results showing distribution and basal expression of O. javanicus actb mRNAs in adult tissues (A) and developing embryos (B). Means with different letters are significantly different based on the ANOVA followed by Duncan’s multiple range tests at P < 0.05. Br, brain; Ey, eye; Fi, fin; Gi, gill; He, heart; In, intestine; Ki, kidney; L, liver; Mu, muscle; Sp, spleen; JF, fertilized; 8C; eight cells; Mo, morula; Bl, blastula; Ga, gastrula; Ne, neurula; 4S, four-somite stage; HB, heart beating stage; RP, retinal pigmentation stage; GBV, gill blood vessel formation stage; VBV, visceral blood vessel formation stage; SD, spleen development stage; JH, just-hatching.
moderate or weak expression (P < 0.05). Our data concur with previous studies on the ubiquitous expression of actb transcripts in a wide array of tissues with variable basal expression levels among tissue types. We showed that the pattern of actb expression transcripts in O. javanicus tissues agreed in general with those from other fish species in terms of the relatively high expression in kidney, intestine, and heart, and low expression in skeletal muscle and liver (Kim et al., 2008; Lee et al., 2009).
The actb gene was developmentally regulated as determined by real-time RT-PCR (Fig. 2B). The actb transcripts were detectable in all of the embryonic samples examined, showing mRNA levels gradually increasing with progression of embryonic development until the blastula stage (P < 0.05). Then the mRNA levels increased sharply, reaching their highest expression at the heartbeat stage (P < 0.05). Then actb expression decreased until the formation of the visceral blood vessels (P < 0.05), after which it remained constant until hatching (P > 0.05). Although the regulation of the actb gene during development is not yet fully understood, the expression pattern observed in O. javanicus is similar not only to that in the related ricefish, O. dancena (Cho et al., 2011), but also to distantly related teleost species, the Atlantic halibut Hippoglossus hippoglossus (Fernandes et al., 2008) and the European seabass Dicentrarchus labrax (Mitter et al., 2009).
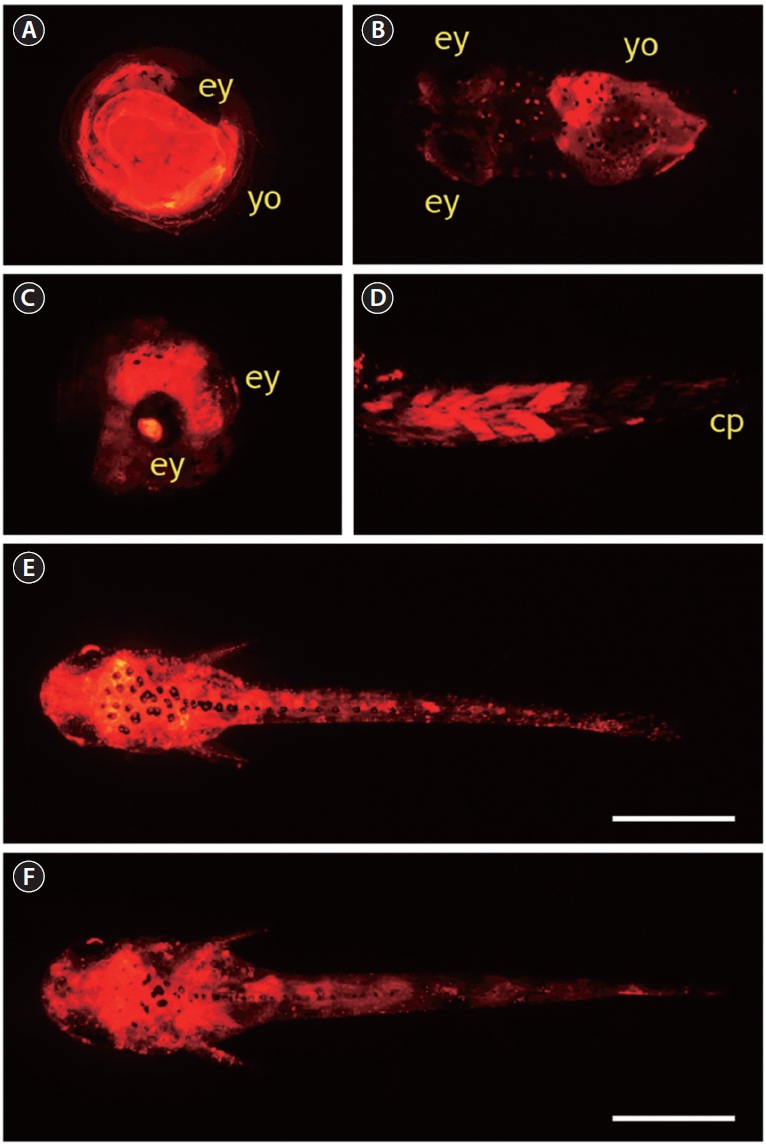
A total of 795 one-celled embryos were injected with pOJβ-actRFP in two independent microinjection trials. The average viability of injected embryos during early development until the gastrula stage was 52%, and the viability from gastrula to hatching was 61%, resulting in an average 30% hatching success, which was significantly lower than the hatchability of the non-injected control group (85%). In the pOJβ-actRFPmicroinjected group, the onset of RFP expression was observed in less than 5% of the embryos at the neurula stage; however, the expression strength was not very high at this stage. When the embryos reached the somite formation stage, about 30% of the surviving embryos started to exhibit active RFP signals, and RFP intensity became stronger as development progressed. In general, these findings concur with those from previous studies on microinjection of the actb promoter-driven transgene construct, although onset of expression of exogenously introduced genes are reported to differ among fish species (Gibbs and Schmale, 2000; Cho et al., 2011). RFP signal distribution was not tissue-specific, as RFP signals were detectable at many sites, including the yolk and embryonic body, as expected from the ubiquitous nature of the cytoskeletal actin gene. Although all of the RFP-positive embryos had a mosaic RFP signal distribution, the expression patterns in pOJβ-actRFP-microinjected embryos could be broadly categorized into three groups. First, more than 50% of RFP-positive embryos displayed RFP expression only in very restricted area(s) of the embryonic body or the yolk. This could be attributed to the mosaic status of the transgene, which is a common phenomenon seen in most microinjection-based gene transfer studies in fish embryos (Nam et al., 1999; Hackett and Alvarez, 2000; Cho et al., 2011). Second, a considerable number of embryos showed significant expression of RFP in the yolk without any apparent RFP signals in the embryonic body (Fig. 3A and 3B). This yolk-exclusive (or dominant) pattern could be explained by the crucial roles of cytoskeletal actin proteins in the yolk syncytial layer (YSL) epiboly during fish embryogenesis (Carvalho and Heisenberg, 2010). YSL is a transient embryonic syncytial tissue, which persists until the larval stage, and it undergoes dynamic movements that are important in morphogenesis and conformational changes in the developing embryos (Carvalho and Heisenberg, 2010). Third, a few embryos (less than 10% of RFP-positive embryos) had a ubiquitous distribution of high RFP expression in almost the entire embryonic body, including the lens of the eye (Fig. 3C),
which is a known pattern of actb gene expression, suggesting that the integration of the microinjected DNA occurs in early cleavage, and therefore persists in subsequent cell divisions (Cho et al., 2011). In several hatchlings, strong RFP expression in the muscles was observed, especially in dorsal and peduncle muscles (Fig. 3D), which was not consistent with the low expression of endogenous actb transcripts in skeletal muscle of adult fish. This suggests that the transcriptional regulation of the actb gene in muscles might differ between early larvae and adults. Several RFP-positive larvae developed from pOJβ-actRFP-microinjected embryos displayed a wide distribution of RFP signals over nearly their entire body (Fig. 3E and 3F), potentially resembling the expression pattern of the endogenous actb gene.
To summarize, we characterized the genetic determinant of O. javanicus β-ACT and evaluated its 5´-upstream regulatory region, using microinjected embryos in heterologous expression assays. O. javanicus β-ACT shares conserved features with its vertebrate orthologues at both the nucleotide and amino acid levels. The actb gene transcripts were widely distributed in all adult tissues with differential levels of basal expression. During development, the O. javanicus actb mRNA was also differentially modulated. In embryos microinjected with the O. javanicus actb gene promoter-driven RFP reporter vector, a ubiquitous distribution of RFP signals was observed, although all of the embryos and resultant hatchlings did not have a uniform expression due to the mosaic nature of the introduced transgene. Data from this study could provide a useful basis on which to develop a transgenic platform representing strong and ubiquitous expression of various foreign genes in O. javanicus.