Coastal or marine environments are often defined as the “final destination” for pollutants that arise from land-based anthropogenic and industrial activities, and this means that the living organisms that inhabit coastal areas are often chronically exposed to various pollutants (Koh et al., 2005; Aoki et al., 2010). Among these pollutants, xenoestrogens are potent endocrine-disrupting chemicals (EDCs) that deregulate gene expression and enzymatic functions associated with the reproduction of the exposed organisms (Rotchell and Ostrander, 2003; Tokumoto et al., 2004, 2011). Previous studies have proposed that exposure to EDCs results in adverse effects on other physiologic attributes, including the innate immunity and host defense mechanisms, of the exposed animals (Lam et al., 2011; Brander et al., 2012). For this reason, understanding the gene expression pattern following EDC exposure is important for developing effective risk management strategies in relation to EDC-caused pollution in marine and coastal waters.
Fish are considered to be a useful model system for studying the ecotoxicology and ecotoxicogenomics of aquatic and marine environments, particularly for addressing EDC-related problems, as a fish biomarker approach can be used to assess the bioavailability, bioaccumulation, toxicokinetics, and meta-bolic conversion of EDCs (Scholz and Mayer, 2008; Jin et al., 2011). These assessments are often not feasible with either traditional analytical chemistry-based assays or in vitro assays using cultured cells, which may fail to correlate in vitro effects to effects on whole organisms (Kurauchi et al., 2005). Based on these advantages, various estrogen-responsive genes from fish have been exploited as potential biomarkers to detect waterborne EDCs (Lee et al., 2002a; Rotchell and Ostrander, 2003; Williams et al., 2007).
Choriogenins are egg envelope proteins that are mainly delivered from the livers of matured female fish (Lee et al., 2002a; Chen et al., 2008; Hong et al., 2009). As the major protein constituents of the thick inner layer of the egg envelope, choriogenin H (ChgH) is an isoform with a molecular mass of about 75 kDa, while its counterpart isoform, choriogenin L (ChgL), has a molecular mass of about 45 kDa. It is known that fish ChgH (as well as ChgL) is synthesized in the liver in response to estrogen signaling (i.e., during the maturation of females) and thereafter transported to the ovary (Lee et al., 2002b; Ueno et al., 2004; Fujita et al., 2008). Although this egg envelope protein isoform was originally considered to be a female-specific gene product, more recent studies have indicated that the transcription of choriogenin genes is detectable and inducible in male livers, and even in late-stage embryos (Yu et al., 2006). Given its high-level inducibility by various estrogenic chemicals, many previous studies have suggested the application of choriogenin H gene expression as an estrogen- sensitive biomarker for aquatic and marine ecotoxicology (Lee et al., 2002a; Chen et al., 2008).
Marine medaka Oryzias dancena, Beloniformes is closely related to a widely used popular model fish, Japanese medaka O. latipes. However, unlike the freshwater-originating O. latipes, the marine medaka is a truly euryhaline species with significant capacities for hypo- and hyper-osmoregulation (Song et al., 2009; Cho et al., 2010). The marine medaka display similar development, growth, and reproduction under various salinity conditions, ranging from complete freshwater to normal seawater (Cho et al., 2010, 2011). This attractive feature enables this euryhaline species to be used as a versatile in vivo biological test system for freshwater, brackish water, and seawater environments. In line with our long-term goal to develop this marine medaka as an ecotoxicogenomic model for marine and aquatic environments, in the present study, we characterized the genetic determinants of choriogenin H in marine medaka. For this purpose, we analyzed the odChgH gene at the transcript and genomic levels, characterized its 5′-flanking regions, and examined the basal- and E2-induced expression profiles of the odChgH transcripts.
The marine medaka O. dancena specimens used in this study were a laboratory-propagated strain maintained in the Institute of Marine Living Organisms, Pukyong National University. Details of the strain characteristics can be found elsewhere (Song et al., 2009). Unless specified otherwise, the fish were maintained throughout the lifecycle at 25℃ at a salinity level of 10 ppt. The fish were fed an artificial diet designed for flounder larvae (Woosung Feed Corp., Daejeon, Korea).
To isolate the structural gene for odChgH, the PCR primers ORY-ChgH G1F and ORY-ChgH G1R, which are conserved to known choriogenin H genes from Oryzias species, were used. All the PCR primers used in the present study are listed in Table 1. The amplified genomic fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and six randomly chosen recombinant clones were sequenced in both directions. The genomic segment that included the 5′-flanking region of odChgH was PCR-amplified using the primers: ORY-ChgH P1F (conserved among Oryzias species); and ODChgH I1R (complementary to intron 1 of odChgH). The amplified product was TA-cloned and sequenced, as above. To obtain the full length 3′-untranslated region (UTR), genome walking was performed, as several degenerate primers proved unsuitable. Designed genome walking primers (ODChgH GW1F and ODChgH GW2F) were paired with adaptor primers (AP1 and AP2; provided in the Universal GenomeWalker Kit; Clontech Laboratories, Mountain View, CA, USA). Genome walker libraries were generated by restriction digestion of marine medaka genomic DNA with DraI, EcoRV,
PvuII, and StuI. All the procedures for preparation of the PCR template and thermal cycling were carried out according to the manufacturer’s instructions. The amplified fragments were TA-cloned and sequenced. Finally, to isolate a continuous version of odChgH, the genomic fragment that spans from the 5′-flanking region to the 3′-UTR was PCR-amplified using the primer pair of ODChgH-FW and ODChgH-RV, TA-cloned, and sequenced.
To generate the cDNA copy of odChgH gene, vectorette PCR was carried out using an excised phagemid stock of marine medaka whole-body cDNA library (unpublished data). The gene-specific primer ODChgHc1F was paired with a T7 vector primer, the binding site of which was located at the end of a multiple cloning site in the pBluescript SK(-) phagemid vector used for the construction of the marine medaka cDNA library (Stratagene Product Division, Agilent Technologies, La Jolla, CA, USA). The fragment amplified by vectorette PCR was TA-cloned and sequenced. Based on the preliminary sequence, the full-length odChgH cDNA was PCR-amplified using the primers ODChgHc 1F and ODChgHc RV, and subsequently sequenced.
The structure and organization of odChgH were compared with those of the corresponding orthologues available in GenBank (http://www.ncbi.nlm.nih.gov) and/or the Ensembl genome database (http://www.ensembl.org). Predictions of potential zona pellucida (ZP)/Trefoil factor family (TFF) domains and phosphorylation sites in the deduced ChgH polypeptide sequence were made using the ExPASy Server (http://prosite.expasy.org). A potential cleavage site for the signal peptide was determined using the SignalP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP/). Transcription factor (TF) binding sites or motifs in the 5′-flanking region were predicted with the TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME) and TFSEARCH tools (http://www.cbrc.jp/research/db/TFSEARCH.html).
To examine the basal expression levels of ChgH transcripts in adult tissues under the non-exposed condition, nine tissues (brain, gill, heart, intestine, kidney, liver, skeletal muscle, ovary, and testis) were surgically obtained from eight females and eight males (9-month-old fish; average total length, 3.8 ± 0.4 cm), respectively. Total RNA was extracted using the TriPure reagent (Roche Applied Science, Mannheim, Germany), followed by purification with the RNeasy Mini Kit, including the DNase treatment step using the RNase-free DNase Set (Qiagen, Hilden, Germany). An aliquot (2 μg) of total RNA was reverse-transcribed into cDNA using the Omniscript RT-Kit (Qiagen) according to the manufacturer’s instruction. During the reverse transcription (RT) reaction, an O. dancena 18S rRNA reverse primer (OD18S RV) was also included, so as to prepare the normalization control for input total RNA across the samples (Cho et al., 2011). Based on preliminary tests, end-point, semi-quantitative RT-PCR (initial denaturation step of 94℃ for 2 min, followed by 24 cycles of 94℃ for 20 s, 58℃ for 20 s, and 72℃ for 20 s) was performed with the qODChgH FW2 and qODChgH RV2 primers. The normalization control of 18S rRNA was also amplified from each sample using the qOD18S-1F/1R primers under the thermal cycling conditions described above, albeit for only 20 cycles. The amplified product was visualized using ethidium bromide staining and analyzed with the Quantity-One image analysis software implemented in VersaDoc 4000 (Bio-Rad, Hercules, CA, USA).
To examine the developmental stage-related expression of ChgH transcripts under the non-exposed condition, embryonic and larval samples were obtained. Embryos (>24 embryos each) were incubated at 25℃ in 10 ppt salinity and collected at the following developmental stages: 1) just-fertilized; 2) 4-cell; 3) 32-cell; 4) morula; 5) blastula; 6) gastrula; 7) neurula; 8) 4-somite; 9) formation of tubular heart; 10) heartbeating; 11) onset of blood circulation; 12) retinal pigmentation; 13) gill blood vessel formation; 14) formation of visceral blood vessels; 15) spleen development; and 16) just-hatching. Details of the embryonic development stages are described in our previous publication (Song et al., 2009). Total RNA was isolated using the Ribospin Total RNA Kit (GeneAll, Seoul, Korea). Preparation of cDNA and RT-PCR assays were conducted with the primers described above.
The preliminary tests and basal expression assay conducted in the present study showed that: 1) female marine medaka adults already displayed very high levels of odChgH mRNA in multiple tissues; and 2) spawning females often showed large individual variations in the basal level of odChgH expression. For these reasons, we employed only male adults in the assays of ChgH expression in response to estradiol-17β (E2) exposure. First, to select for the expression assay appropriate candidate tissues that undergo potent induction of odChgH transcripts in response to E2, eight males (8-month-old fish; average total length, 3.8 ± 0.3 cm) were immersed in a 10-L tank that contained 8 L of well-aerated water supplemented with E2 (1.0 μg/L). The salinity was adjusted to 15 ppt using a synthetic salt (Kent Marine, Acworth, GA, USA) and the temperature was maintained at 25.0 ± 0.5℃ throughout the exposure. Treatment duration was fixed at 5 days. A non-exposed control group of similar-sized males (n = 8) was prepared in an identical manner, except that E2 was not added to the tank. The fish were fed an artificial diet for flounder larvae (500 μm in diameter; see above) once daily. Half of the water in the tank was exchanged on Days 2, 4 and 6 post-exposure, and the hormone treatment was renewed for the E2-exposed group. When the treatment was finished, selected tissues (brain, eye, fin, gill, heart, intestine, kidney, liver, skeletal muscles, spleen, and testis) were obtained individually from each male in the E2-exposed group and non-exposed group. Semi-quantitative RT-PCR was performed as described above, to identify the most sensitive tissues in terms of odChgH induction by E2 exposure. Second, using the tissues (liver and kidney) selected on the basis of the first exposure experiment (above), dosedependent induction of odChgH transcripts was carried out using a series of E2 doses (0.1, 0.3, 0.5, 0.7, 0.9 μg/L). Male adults (n = 6/tank) were allocated to one of five tanks (10-L) that contained water at 15 ppt salinity and E2 at the desired concentration, and the fish were exposed to the hormone for 1 week. A non-exposed control group was prepared in the same manner, except that no hormone was added. The other experimental conditions were the same as those described above. After exposure to E2, tissues were sampled for quantitative Real-time RT-PCR of odChgH transcripts (see below). Third, the effects of different environmental salinity levels on the efficiency of induction of odChgH transcripts by E2 were examined. Adult males (n = 10) were pre-acclimated to freshwater (salinity, 0 ppt), brackish water (15 ppt), and seawater (30 ppt) for 2 weeks. After the pre-acclimation period, five fish from each salinity group were exposed to E2 (0.3 μg/L), while the remaining five fish were prepared as non-exposed controls. Treatment duration was 1 week, and the other treatment conditions were the same as described above. The levels of odChgH transcripts were assayed by real-time quantitative RT-PCR (qRT-PCR).
The total RNA, cDNA template, and PCR primer pairs were prepared as described above for the end-point RT-PCR assay. The half-diluted cDNA template was subjected to PCR (an initial denaturation step of 94℃ for 2 min, followed by 45 cycles of 94℃ for 20 s, 58℃ for 20 s, and 72℃ for 20 s) with the iQ™ SYBR Green Supermix (Bio-Rad) and the iCycler Real-Time Detection System (Bio-Rad). Based on the standard curve for each gene (odChgH and 18S rRNA) prepared using a serial dilution of the positive plasmid, the average PCR efficiencies for odChgH and 18S rRNA were 97.0% and 95.0%, respectively. Differential expression of odChgH in the E2-exposed groups relative to the non-exposed control groups are expressed as fold-changes using the following formula: relative expression = (1 + EChgH)ΔCtChgH/(1 + E18S rRNA) ΔCt18SrRNA, where ΔCt is the Ct non-exposed - Ct E2-exposed (Schmittgen and Livak, 2008). Triplicate assays for each cDNA sample were performed in an independent manner. Differences between or among samples were assessed with the Student’s t-test or ANOVA, followed by Duncan’s multiple range test at a P-value of 0.05.
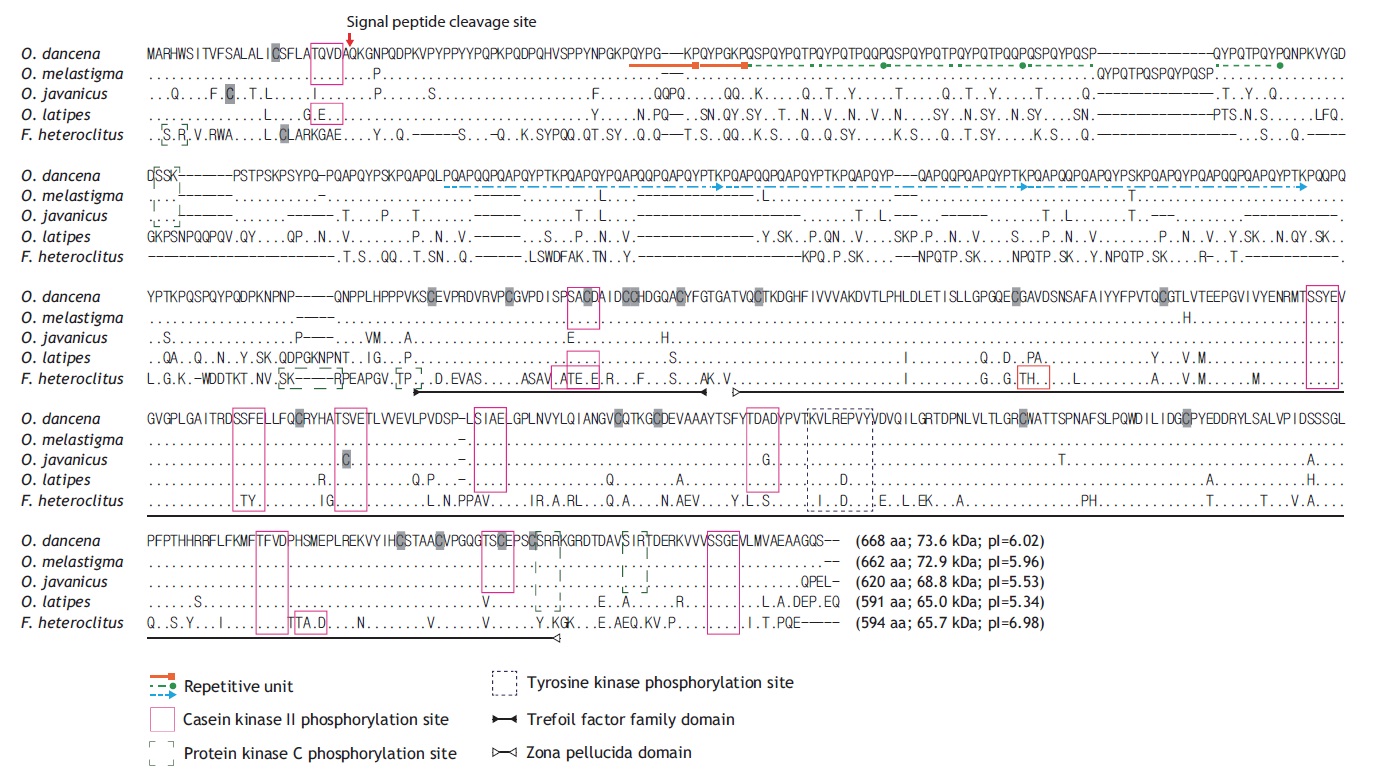
The full-length cDNA (2,135 bp) contains an 18-bp 5′- UTR, an open reading frame (ORF; 2,004-bp excluding the stop codon) that encodes the ChgH polypeptide of 668 amino acids (aa), and a 113-bp 3′-UTR, which includes the poly(A)+ tail (GenBank accession no. JQ268272). A predicted polyadenylation signal ATTAAA was located 17 bp before the poly(A)+ tail. The deduced ChgH polypeptide (molecular mass of 73.6 kDa, with a theoretical pI value of 6.02) shows a predicted signal peptide cleavage site between Ala26 and Gln27. Multiple sequence alignments with the ClustalW algorithm indicate high sequence similarity with other teleostean orthologues at the amino acid level, particularly with those from related species of the same Oryzias genus (Fig. 1). The O. dancena ChgH shares characteristics typical of fish ChgH proteins with representative orthologues, including the conserved phosphorylation sites and TFF/ZP domains. As is the case for other ChgH orthologues, the marine medaka ChgH possesses a long, proline- and glutamine-rich , repetitive N-terminal region, in which the repetitive patterns are: (QYPGKP)2, [QSPQYPQ(T/S)PQYPQTPQ(Q/Y)P]3 and [PQAPQQPQAPQYP(T/S)KPQAPQYPQAPQQPQAPQYPTK] 3. However, the repetitive units and their copy numbers observed in the published ChgH sequences show variability, even among Oryzias species.
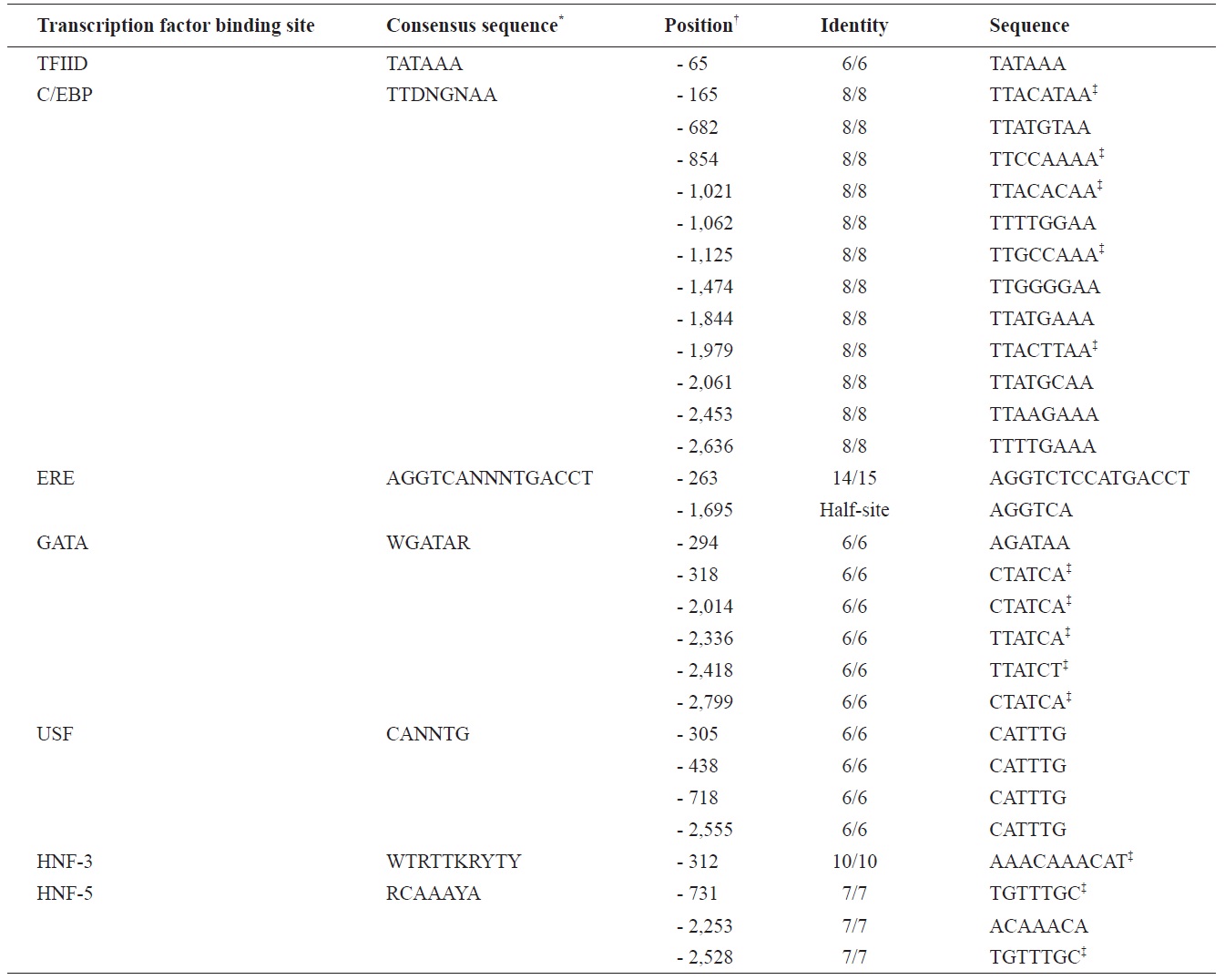
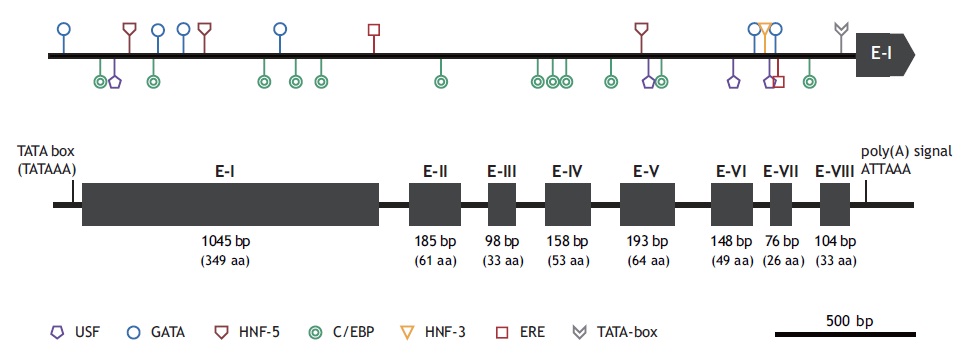
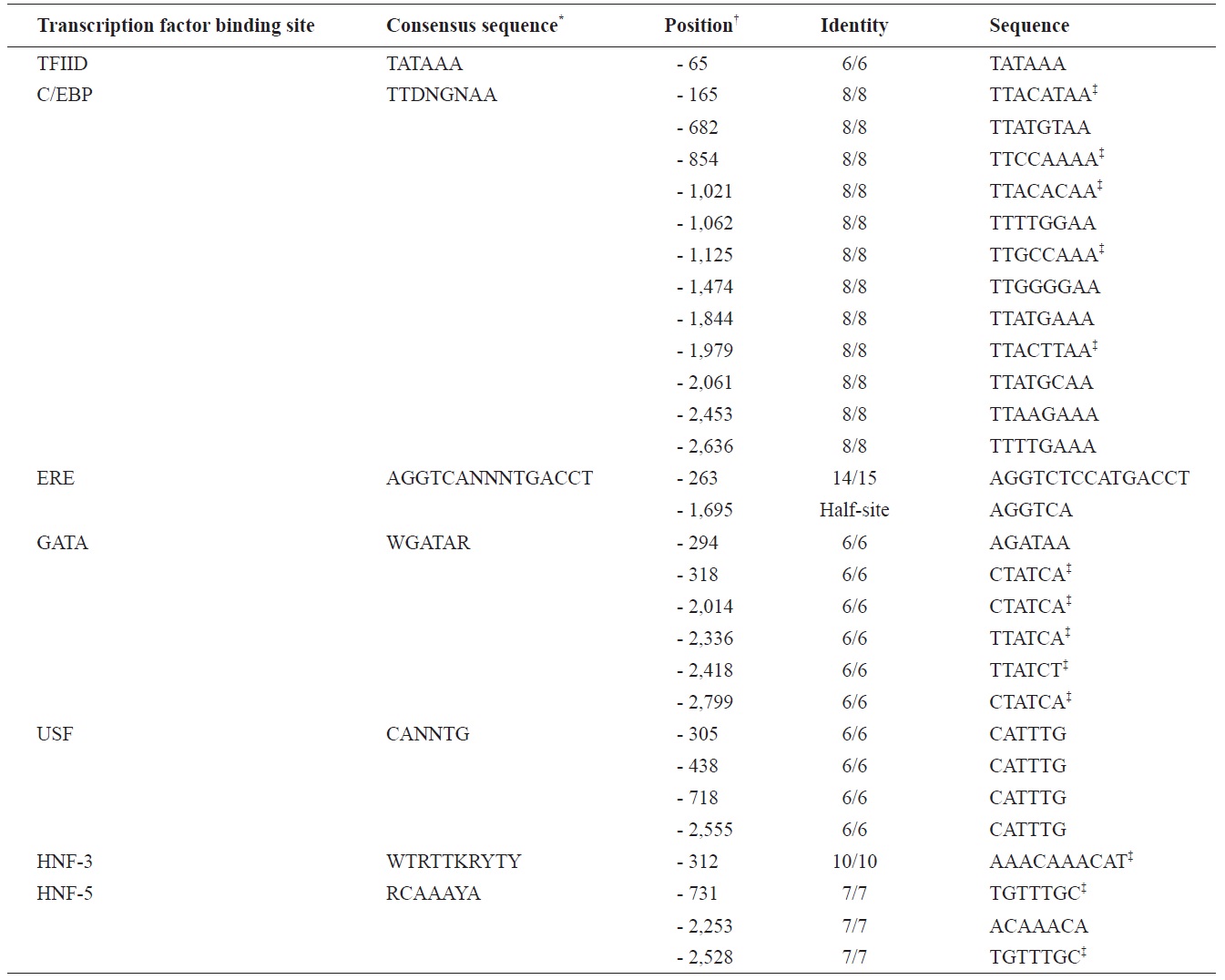
The marine medaka ChgH gene (odChgH; GenBank accession no. JQ268271) possesses eight exons (1,045, 185, 98, 158, 193, 148, 76, and 104 bp for exon I to exon VIII, respectively) interrupted by seven introns (90, 90, 112, 86, 136, 62, and 93 bp for intron I to intron VII, respectively) within a 2.67-kb region that spans from an ATG translation initiation codon to a stop codon (Fig. 2). The coding regions of the genomic odChgH were clearly matched to its cDNA counterpart. The AG/GT exon-intron boundary rule was consistently found at each junction. Based on the bioinformatics prediction, the 2,832-bp 5′-UTR of odChgH was proven to possess various TF binding motifs/elements, as well as a canonical TATA box (TATAAA; -65 bp from the ATG site) (Table 2, Fig. 2). These elements include the GATA box (consensus sequence: WGATAR; 6 copies), CCAAT/enhancer binding site (C/EBP:
TTDNGNAA; 12 copies), upstream stimulatory factor site (USF: CANNTG; 4 copies), hepatocyte nuclear factor binding site (HNF: RCAAAYA or WTRTTKRYTY; 4 copies) and estrogen response element (ERE: AGGTCANNNTGACCT; 1 copy). For ERE, one additional half site (AGGTCA) was predicted at -1,695 bp.
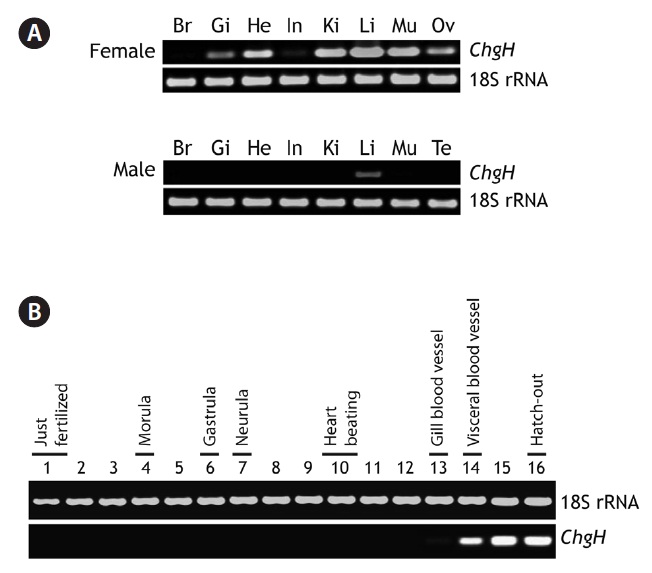
Marine medaka female and male adults exhibited different basal expression patterns of odChgH transcripts under nonexposed conditions (Fig. 3A). For the females, odChgH transcripts were detected in a wide array of tissues, including the
gills, heart, intestine, kidney, liver, muscle, and ovary, and the basal mRNA levels in these tissues, as assessed by RT-PCR, were variable. Although odChgH transcripts predominated in the female liver, the levels of odChgH mRNA in other female tissues, such as kidney and muscle, were comparable to those found in the liver. In addition to the robust expression of odChgH mRNA observed in these three tissues, moderate levels of odChgH expression were detected in the gills, heart, and ovary, while the intestine exhibited only weak expression of this gene. In contrast, odChgH transcripts were not detected in the brain using our RT-PCR conditions. Moreover, unlike the females, odChgH mRNA in the males was observed exclusively in the liver. The basal expression level of hepatic odChgH transcripts in the male was much lower than that in the female liver.
During embryonic development, the expression of odChgH mRNA was scarcely detectable until the gill blood vessel formation stage (4 days and 20 h post-fertilization). Even at this stage, the expression level was rather low; only a faint amplification signal was seen with extended PCR cycles. However, when the embryos reached the stage of visceral blood vessel formation (6 days and 12 h post-fertilization), odChgH expression was significantly increased. Thereafter, the expression level increased at the heart and spleen development stage (8 days post-fertilization) and stabilized in the just-hatched larvae (11 days post-fertilization) (Fig. 3B).
[Fig. 3.] Representative reverse transcription (RT)-PCR gels showing marine medaka Oryzias dancena choriogenin H (ChgH) transcripts expressed in adult tissues (A) and developing embryos (B). RT-PCR bands of 18S rRNA are also shown as the normalization control. Br, brain; Gi, gill; He, heart; In, intestine; Ki, kidney; Li, liver; Mu, skeletal muscle; Ov, ovary; Te, testis. In (B), the lanes from 1 to 16 indicate the important developmental stages of O. dancena based on the Song et al. (2009).
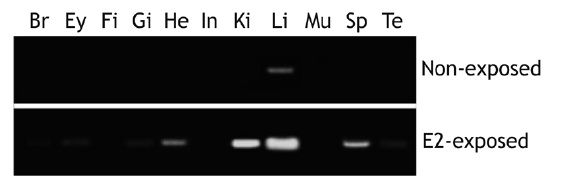
[Fig. 4.] Induction of choriogenin H (ChgH) mRNAs in different tissues marine medaka Oryzias dancena males by exposure to estradiol-17β (E2; 1 μg/L for 5 days). Br, brain; Ey, eye; Fi, fin; Gi, gill; He, heart; In, intestine; Ki, kidney; Li, liver; Mu, skeletal muscle; Sp, spleen; Te, testis.
After exposure to E2 (1.0 μg/L for 5 days), marine medaka males showed differential induction of odChgH transcripts, with the level of induction being dependent upon the tissue type (Fig. 4). Although control males (non-exposed) had negligible levels of odChgH transcripts in their non-liver tissues, E2 exposure resulted in significant elevation of odChgH transcripts in various non-liver tissues, which was readily detectable by RT-PCR. While the liver showed the strongest induction of odChgH mRNA, several tissues, including the heart, kidney, and spleen, showed the amplification products of odChgH mRNA. In terms of the end-point level of induced odChgH mRNA, the liver had a higher level than the
other tissues. Induction was highest in the liver, followed by the kidney, spleen, and heart, while the brain, eyes, gills, and testis showed weak induction of odChgH transcripts. The de novo induction of ChgH mRNA by E2 in the fin, intestine, and skeletal muscle was weak, as detected using our RT-PCR conditions.
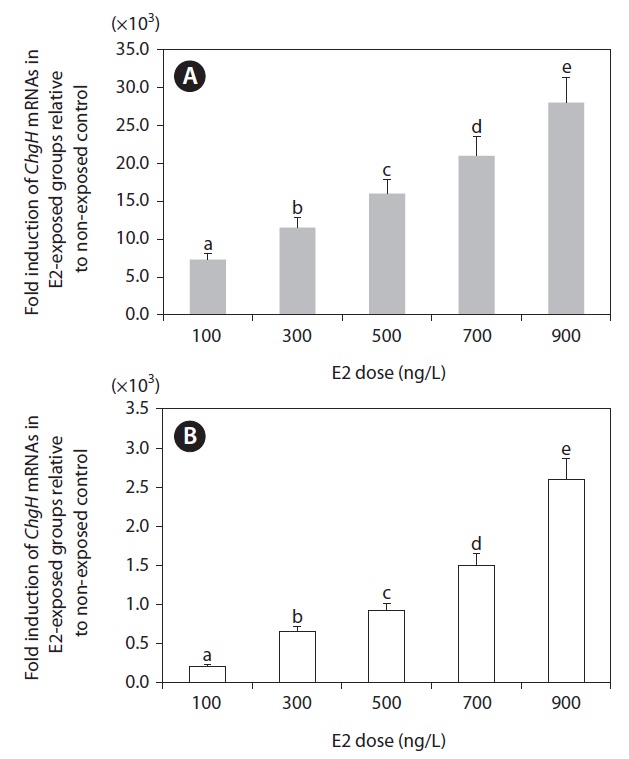
Based on the tissue expression assays described above, we selected the two most sensitive tissues (liver and kidney) for further analysis. Although the spleen showed comparable E2 sensitivity, spleen samples sufficient for repetitive RT-PCR assays were lacking. As shown in Fig. 5, the transcriptional response of odChgH to E2 was dependent upon the dosage of E2, whereby there was a clearly positive relationship between the induced odChgH mRNA levels and E2 dosages. In the livers of the E2-exposed fish, the fold-induction values for odChgH mRNA relative to the levels in the livers of the non-exposed controls were 7,300, 11,500, 16,000, 21,000, and 28,000, respectively, for fish exposed to 0.1, 0.3, 0.5, 0.7, and
0.9 μg/L E2. In contrast, the induction levels for the kidneys ranged from 210-fold (0.1 μg/L E2) to 2,600-fold (0.9 μg/L E2) relative to the basal expression observed in non-exposed control fish.
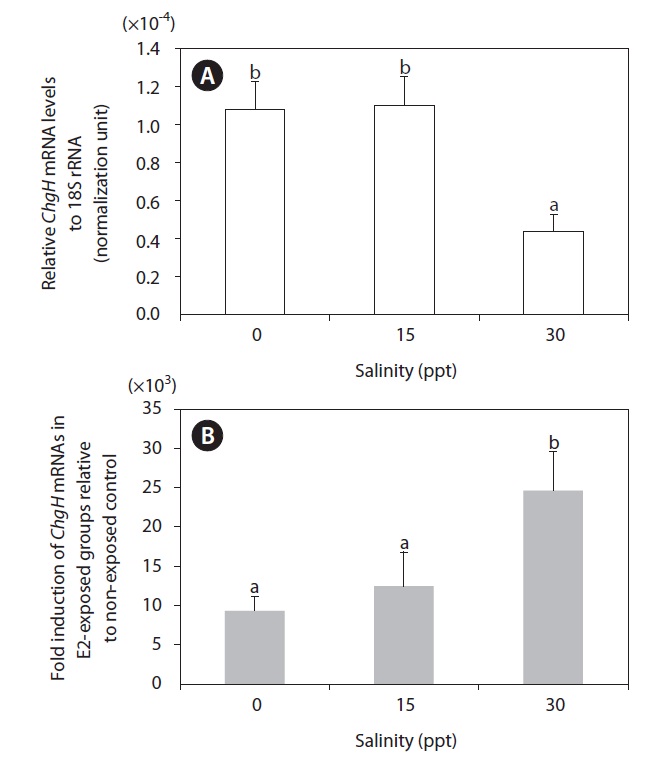
Induction of hepatic odChgH transcripts by E2 exposure occurred at all the salinity levels tested (Fig. 6). The fold-induction for the E2-exposed group relative to the non-exposed group differed depending on the environmental salinity level, whereby induction was strongest in seawater (30 ppt; >2.40 × 104-fold, as compared to the basal level) (P < 0.05). The level of induction in brackish water (15 ppt; 1.21 × 104-fold) was slightly higher than that in freshwater (0 ppt; 9.31 × 103-fold), although the difference between the groups was not statistically significant due to the large variation among E2-exposed individuals in the brackish water (P > 0.05). However, the large differences in fold-induction of odChgH mRNA between the 30-ppt and the 0-ppt and 15-ppt groups was closely correlated with the differential basal levels of odChgH mRNA among the salinity groups. The initial expression levels of hepatic odChgH mRNA in non-exposed marine medaka males were markedly lower at 30 ppt (average, 4.39 × 10-5; normalization against 18S rRNA) than at 0 ppt (1.08 × 10-4) and 15 ppt (1.10 × 10-4) (P < 0.05).
In the present study, the gene and promoter structures for choriogenin H were determined in a marine medaka species O. dancena, and the mRNA expression patterns of this gene under both non-induced and E2-induced conditions were examined. The odChgH shares typical structural features with other fish orthologues at both the mRNA and deduced amino acid levels. The conserved characteristics include: the 1) proline- and glutamine-rich N-terminal region (Murata et al., 1997); 2) the presence of TFF and ZP domains, which contain multiple cysteine residues that form intramolecular disulfide bonds, which are of importance for the structural integrity of the ChgH protein (Kanamori et al., 2003; Yu et al., 2006; Fujita et al., 2008); 3) conserved phosphorylation sites that are targeted by casein kinase II, protein kinase C, and tyrosine kinase (Yu et al., 2006; Chen et al., 2008); and 4) a conserved cleavage site for the signal peptide. As shown in the alignment of the protein sequences (see Fig. 1), the species-specific differences among the ChgH proteins are mainly in the repetitive N-terminal domain, which suggests that this domain has experienced a more rapid evolution than other parts of the protein in teleosts (Arukwe and Goksøyr, 2003). This Nterminal domain is postulated to be involved in the hardening process in fertilized eggs by forming isopeptide bonds, and it is proposed that the degree of repetitiveness is related to the differential hardness of chorions among fish species (Wang and Gong, 1999; Liu et al., 2006; Chen et al., 2008). Overall, the homology pattern observed for marine medaka ChgH was in agreement with the previously known taxonomic appraisal of the species examined in that the present ChgH showed the highest similarity with that of another euryhaline medaka species, O. melastigma (Chen et al., 2008). Previous studies have proposed that O. dancena be designated as the same species as O. melastigma, based mainly on their morphologic and euryhaline characteristics (Inoue and Takei, 2002, 2003). However, the odChgH isolated in the present study is not identical to that previously reported for O. melastigma. Pairwise alignment of the two protein sequences (from O. dancena and O. melastigma) indicates that there are four amino acid substitutions and a 15-aa deletion/insertion (QYPQTPQSPQYPQSP) in the repetitive domain. In addition to these changes in the repetitive domain, another amino acid substitution (Leu ↔ His) occurs in the region corresponding to exon II. Although it remains unclear as to whether this considerable difference between the two choriogenin H sequences can be regarded in terms of allelic or isoform differences, further careful revision or reconsideration may be needed for the taxonomy and possible subspeciation of O. dancena and O. melastigma.
At the genomic level, odChgH represents a genomic organization that is conserved with other orthologues (Fig. 2). Similar to the other medaka species, odChgH is characterized by eight exons and seven introns. However, it is noteworthy that the second isoform of the ChgH gene, which has only seven exons, appears in the O. latipes genome (Lee et al., 2002a; Kanamori et al., 2003). This suggests that further exploration of the marine medaka genome for the presence of other isoforms is needed. From the bioinformatics analysis of the 5′-flanking region of odChgH, various motifs and elements targeted by different TFs can be predicted. Of the various TF binding sites, ERE (identified in complete or half-length copy) is known to play crucial roles in the estrogen-mediated regulation of many other estrogen-responsive genes (Ueno et al., 2004; Yu et al., 2006). The GATA factor (predicted to be in multiple copies) has been reported to be involved in the transcriptional activation of ERE-mediated gene regulation (Yu et al., 2006). However, the USF site (E-box) has also been commonly found in ZP-related genes (Wilson and Roesler, 2002). The presence of multiple copies of hepatocyte-rich TFs, such as C/EBP and HNFs, is in accord with the liver-predominant expression of ChgH genes in fish (Cardinaux et al., 1994; Schrem et al., 2004; Huang et al., 2010). Further functional studies to gain insights into the coordinated or orchestrated functions of the predicted TFs are needed.
Although choriogenin H was originally reported as a liver- delivered, spawning female-specific protein, more recent studies have indicated that ChgH gene transcription also occurs in the livers of adult males (Lee et al., 2002b; Yu et al., 2006). Furthermore, in the present study, the female marine medaka showed a wide distribution of odChgH mRNA among the various tissues, as adjudged by RT-PCR. Certain tissues, including the heart, kidney, and muscles, revealed active transcription of odChgH, the basal mRNA levels of which were readily comparable to that in the liver (see Fig. 3A). Our finding is similar to the previous observation made for Kryptolebias marmoratus (Rhee et al., 2009), although the transcriptional profiles of ChgH in non-liver tissues have not been studied extensively. During embryonic development, the expression of odChgH mRNA first appears at the onset of the retinal pigmentation stage and increases at the visceral blood vessel formation stage (see Fig. 3B). This finding is in accordance with the liver becoming visible under microscopy and the blood circulation being active at these stages (Song et al., 2009). This timing of onset of expression is generally in agreement with earlier observations for other related species (Ueno et al., 2004; Chen et al., 2008; Mourabit et al., 2011).
In the present study, E2 exposure induced odChgH mRNA in several male tissues (e.g., heart, kidney, and spleen) that did not show notable basal expression under the non-exposed condition, which suggests that E2-mediated de novo expression of odChgH occurs in various tissues of the male marine medaka (Fig. 4). Most of the previous postmortem studies have based their results for E2-mediated ChgH expression solely on the observation of liver tissues, with little characterization performed on other tissues (Yu et al., 2006; Chen et al., 2008; Rhee et al., 2009). Therefore, assuming that our observation of de novo induction of ChgH transcripts in non-liver tissues is specific for this medaka species, further extensive studies are warranted.
Based on the results of the qRT-PCR assay, the E2-induced expression of odChgH mRNA was found to be dependent upon the E2 dosage, and there was no saturation of odChgH mRNA induction when E2 was added up to a concentration of 0.9 μg/L (Fig. 5). This dose-dependent induction of odChgH transcription in this species confirms the previous results obtained for O. melastigma males (Chen et al., 2008). However, the fold-increases of hepatic odChgH mRNA in males during E2 exposure observed in the present study (7,300-fold to 28,000- fold with exposure to E2 at 100 ng/L to 900 ng/L, respectively) are higher than those observed previously for E2-exposed O. melastigma males (435-fold at 500 ng/L E2) (Chen et al., 2008). In addition to the liver, odChgH transcription was also induced in male kidneys, suggesting again the possibility that environmental E2 activates the expression of ChgH in many other tissues. However, the fold-induction of odChgH mRNA in the kidney, as determined by qRT-PCR, was not as high as that in the liver. Overall, the data from the present study indicate that quantitative detection of waterborne E2 is possible through the monitoring of odChgH expression in male marine medaka, although further tests need to be conducted in a more environmentally realistic fashion with much lower doses of E2. In terms of the sensitivity to E2 exposure, our data are in agreement with earlier studies showing that ChgH is one of the most sensitive genes to E2 exposure (see below). Yu et al. (2006) and Chen et al. (2008) showed that E2 exposure modulated ChgH expression more sensitively than choriogenin L (ChgL) expression, respectively, in O. javanicus and O. melastigma. Similarly at the protein level, Fujita et al. (2004) observed that ChgH was more responsive than ChgL in the blood serum of E2-exposed masu salmon Oncorhynchus masou.
In the present study, the E2-mediated activation of odChgH mRNA was affected by environmental salinity (Fig. 6). The basal expression of hepatic odChgH mRNA in male marine medaka that were acclimated to seawater (30 ppt) was lower than in those acclimated to either freshwater (0 ppt) or brackish water (15 ppt). In contrast, the inducibility of odChgH mRNA by the same dose of E2 was higher in the fish that were acclimated to seawater than in those acclimated to lower salinity levels. Although several previous studies have reported the successful induction of ChgH mRNA expression following E2 exposure in euryhaline medaka species, the effects of salinity on the E2-mediated expression of ChgH mRNA has received little attention (Yu et al., 2006; Chen et al., 2008). The mechanism underlying this salinity-dependent modulation of ChgH gene expression remains to be elucidated. Moreover, this finding is not in accordance with the previous observation, in the sense that this euryhaline species typically shows similar reproductive characteristics in both freshwater and seawater (Inoue and Takei, 2002; Cho et al., 2010). However, our current data suggest that the lowest-observed-effect concentration of E2 differs for different salinity conditions with the odChgH biomarker. Therefore, studies are needed to clarify the factors that affect the salinity-dependent regulation of odChgH in this euryhaline medaka. In this respect, comparative expression assays for other reproduction-related genes, including relevant transcription factors, in E2-induced and non-induced fish under different salinity conditions would be valuable. In addition, further studies using fish of different ages should be undertaken, since the basal and E2-induced expression levels of ChgH could differ depending on age (or reproductive stage), even in males.
In summary, the genetic determinant for the egg envelope protein isoform choriogenin H was characterized at both the mRNA and genomic levels in marine medaka. The gene in marine medaka possesses conserved structural characteristics typical of fish choriogenin genes. The basal tissue distribution pattern of ChgH mRNA is gender-specific, in that females have higher expression levels and wider tissue distribution patterns than males. Developmentally, the odChgH mRNA appears at the visceral blood vessel formation stage, which is accordance with the degree of liver development in embryos. During E2 exposure in male adults, odChgH mRNA is inducible not only in the liver, but also in other non-liver tissues, although inducibility is highest in the liver. The E2-mediated induction of odChgH mRNA in the liver and kidney is dose-dependent. Finally, both the basal and E2-induced expression levels of ChgH mRNA in male livers are affected by environmental salinity levels, whereby the fold-increase of hepatic odChgH mRNA is significantly higher in seawater-acclimated fish than in brackish water-acclimated or freshwater-acclimated fish.