Genus Echinometra species generally occur throughout the tropical Pacific and also from East Africa to the Indian Ocean (Clark and Rowe, 1971). This genus contains seven extant species, including E. insularis, E. lucunter, E. l. polypora, E. mathaei, E. m. oblonga, E. vanbrunti, and E. viridis (Kroh and Mooi, 2012). Among them, E. mathaei is widely distributed over the Indo-Pacific region from the Red Sea to Hawaii, Australia, and southern Japan (Mortensen, 1943; Nisiyama, 1966). This species was first described by Blainville (1825) based on a single specimen from Mauritius, East Africa. He also described E. m. oblonga as a subspecies of E. mathaei without describing the type locality, and its body color was described as white or violet. Echinometra mathaei oblonga was elevated to the separate genus Mortensia by Doderlein (1906) because of the shape of gonadal spicules. But, Mortensen (1943) claimed these as shapes of E. mathaei, and gave the species a subspecies name, E. mathaei oblonga. Subsequently, Clark and Rowe (1971) and Kroh and Mooi (2012) followed the nomenclature of Mortensen (1943). Some morphological (Arakaki et al., 1998; Arakaki and Uehara, 1999) and molecular (Palumbi et al., 1997; McCartney et al., 2000; Landry et al., 2003) studies were conducted due to the obscure relationship between these species. A morphological and molecular examination and identification of Echinometra specimens collected from Munseom of Jeju Island, Korea was performed in the present study.
Echinoid specimens were collected at depths of 5-10 m in Munseom, Jeju Island by SCUBA diving on November 23, 2008 and September 15, 2009. The specimens were preserved in 95% ethanol, and their important morphological characters were photographed with a digital camera (Canon G12; Canon Co., Tokyo, Japan) and stereo and light microscopy (Nikon SMZ1000, Nikon Eclipse 80i; Nikon Co., Tokyo, Japan). The specimens were morphologically identified based on the descriptions of Mortensen (1943) and Clark and Rowe (1971).
Genomic DNA was extracted from the tubefeet of Korean echinoids using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) for the molecular analysis. The mitochondrial cytochrome c oxidase subunit 1 (COI) gene fragment
was amplified using ECO1a and ECO1b, as suggested by Knott and Wray (2000). The COI gene fragment corresponding to the interval between sites 5921 to 6861 of the Strongylocentrotus purpuratus mitochondrial genome (NCBI ID: NC001453) was obtained for each specimen.
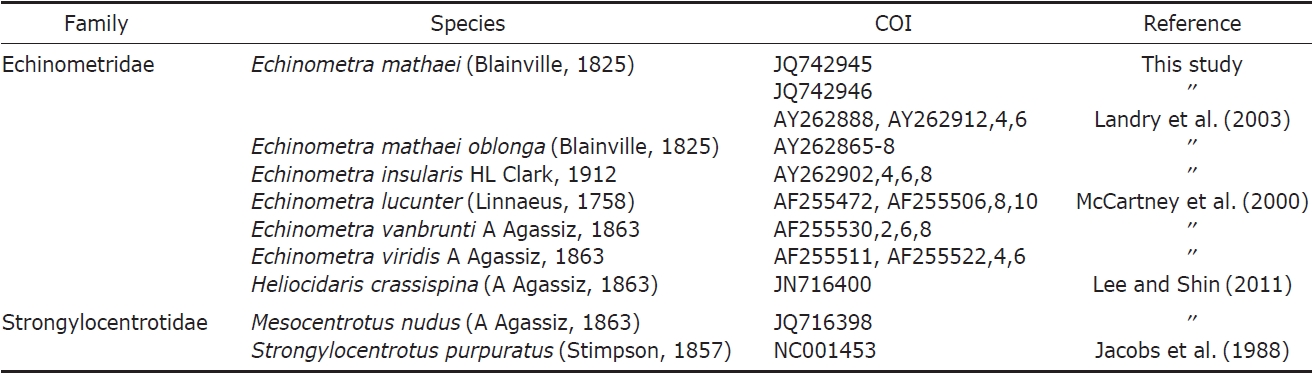
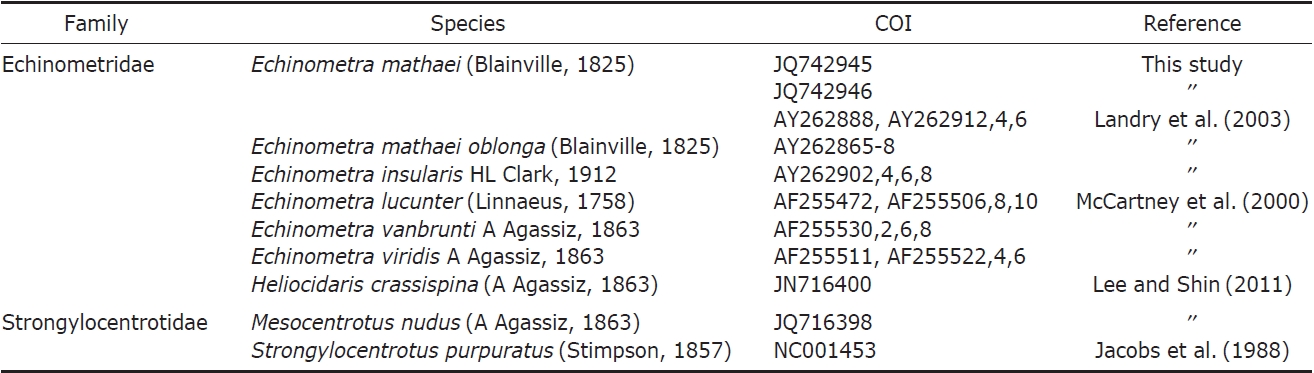
PCR analyses were conducted according to Lee and Shin (2011). DNA fragments were sequenced on an ABI 3730XL Sequencer (Applied Biosystems Inc., Foster City, CA, USA) using the ABI Prism Bigdye Terminator v3.1 (Applied Biosystems). The mitochondrial COI gene was sequenced, and the sequence data of five species that were not distributed in Korea were obtained from GenBank (Table 1). COI sequences were aligned using CLUSTAL X (Thompson et al., 1997), and genetic distances were calculated according to the Kimura 2-parameter model using MEGA5 (Tamura et al., 2011). The best-fit evolutionary model was identified using the Akaike Information Criterion (AIC) in jMODELTEST (Posada, 2008), and the result was GTR+G(α=0.238). A phylogenetic tree was drawn with neighbor joining (NJ), maximum likelihood (ML), and Bayesian inference (BI) methods. The NJ tree was inferred from the Kimura 2-parameter genetic distance and bootstrapped 1,000 times using MEGA5. The ML tree was analyzed with PHYML v.3.0 (Guindon and Gascuel, 2003) and 1,000 bootstrap replications. The BI analysis was conducted with the same model and analyzed by MrBayes 3.1.2 with 2×106 trees, sampling every 1,000th tree, and credibility values of the nodes were calculated with the 50% majority rule tree after discarding the first 500 trees (Ronquist and Huelsenbeck, 2003).
Class Echinoidea Leske, 1778
Order Camarodonta Jackson, 1912
Infraorder Echinidea Kroh and Smith, 2010
Family Echinometridae Gray, 1855
Key to the genera of family Echinometridae in Korea
1. Test oval shaped, with usually four pore-pairs to an arc ????????????????????????????????????????????????????????????????Echinometra
Test nearly circular shaped, with usually eight pore-pairs to an arc ???????????????????????????????????????????????????Heliocidaris
1*Genus Echinometra Gray, 1825
Echinometra Gray, 1825: 426; Blainville, 1830: 206; L Agassiz and Desor, 1846: 372; A Agassiz, 1872-1874: 282; Doderlein, 1906: 233; HL Clark, 1912: 370; Mortensen, 1943: 352; Nisiyama, 1966: 267; Kroh and Mooi, 2012: 179659.
Ellipsechinus Lutken, 1864: 165.
Mortensenia Doderlein, 1906: 233.
Test more or less oval shaped, stout, longitudinal axis through interambulacral III to ambulacral 1. Ambulacrals with four pore-pairs per an arc. Apical system with all exserted ocular plates. Primary spines rather long, strong, pointed, Globiferous pedicellariae with one lateral tooth, tubefeet with C-shaped spicules.
Type species: Echinus lucunter Linnaeus, 1758.
2*Echinometra mathaei (Blainville, 1825) (Fig. 1)
Echinus mathai Blainville, 1825: 94; 1830: 206.
Echinometra mathaei Blainville, 1834: 225; Mortensen, 1903: 128; A Agassiz and HL Clark, 1907: 241; HL Clark, 1908: 303; 1912: 372; Koehler, 1914: 250; Mortensen, 1940: 103; 1943, 381; Nisiyama, 1966: 271; AM Clark and Rowe, 1971: 157; Arakaki et al., 1998: 159; Arakaki and Uehara, 1999: 551; Kroh and Mooi, 2012: 213383.
[Fig. 1.] Echinometra mathaei. A, Dorsal side; B, Ventral side; C, Lateral side; D, Apical system; E, Pore pairs; F, Primary spine on dorsal side (left) and secondary spine on ventral side (right); G, Globiferous pedicellaria; H, Heads of globiferous pedicellaria; I, Valve of tridentate pedicellaria; J, Valve of ophiocephalous pedicellaria; K, Valves of tripyllous pedicellaria; L, Spicules of tubefeet. Scale bars: A-C=1.5 cm, D, F=0.5 cm, E=1.5 mm, G, J=200 μm, H, I=100 μm, K=50 μm, L=25 μm.
Echinometra megastoma M’Clelland, 1840: 181.
Echinometra heteropora L Agassiz and Desor, 1846: 372.
Echinometra microtuberculata A Agassiz, 1863: 22.
Echinometra picta A Agassiz and HL Clark, 1907: 241; HL Clark, 1912: 373.
Ellipsechinus decryi Lambert, 1933: 47.
Material examined. 1 specimen, Munseom, Jeju Island, 23 Nov 2008, at 5 m depth by SCUBA diving; 1 specimen, 15 May 2009, at 10 m depth by SCUBA diving.
Description. Test of small size, strong, hemispherical, oval shaped; ventral side flat, scarcely sunken towards peristome. Ambulacral with four pore-pairs, rarely five to one oblique and rather irregularly curved pore-arcs (Fig. 1E). One of tubercles in pore-zone enlarged and forms a fairly conspicuous vertical. In interambulacral, larger secondary tubercles usually form a distinct vertical series admedially and adradially to primary series. These secondaries vary in size, from almost as large as smaller than primaries. Apical system with all ocular plates exserted (Fig. 1D). Genital plates covered with some small spines, and genital pores developed (Fig. 1D).
Suranal plate indistinct, and anal opening acentric. Primary spines rather long, about as long as half test diameter, usually rather stout, tapering, and colors getting lighter at tips (Fig. 1F). Secondary spines rather short, with flat tips, looks like rolling pins (Fig. 1F). Globiferous pedicellariae very scarce: valve with a lateral tooth as long as two-thirds of apical tooth (Fig. 1H). Tridentate pedicellariae large: valve with curved tips (Fig. 1I). Ophiocephalous pedicellariae narrow in middle: valve with zigzag edges (Fig. 1J). Triphyllous pedicellariae small: valve width moderately as wide as basal part (Fig. 1K). Tubefeet with C-shaped spicules (Fig. 1L). Buccal plates with some small spines in peristome among a few assemblages of ophiocephalous pedicellariae.
Size. Test diameter, 19.0, 29.0 mm; test height, 9.0, 13.0mm (44.8, 47.4% of test diameter); apical system, 3.8, 6.1 mm (18.0, 19.0% of test diameter); peristome, 9.8, 15.0mm(51.6, 51.7% of test diameter); number of interambulacral plates, 9, 13; number of ambulacral plates, 11, 26.
Distribution. Korea (Jeju Island), Japan (Okinawa), China, Philippines, Hawaii, Solomon Islands, northern Australia, Indo-West Pacific, East Africa (Kenya, Mauritius, Mozam-
bique, Tanzania), Red Sea.
The sequences of our Korean E. mathaei specimens were compared with GenBank data of six Echinometra species such as E. mathaei, E. m. oblonga, E. insularis, E. lucunter, E. vanbrunti, and E. viridis of the family Echinometridae, using Heliocidaris crassispina, Hemicentrotus pulcherrimus, and Strongylocentrotus pallidus of the family Strongylocentrotidae within the same infraorder as outgroups taken from GenBank. After sequence alignment, a portion of the COI gene corresponding to bases 6471-6854 of the S. purpuratus complete mitochondrial genome was obtained. Our COI dataset comprised 383 bp of the COI gene.
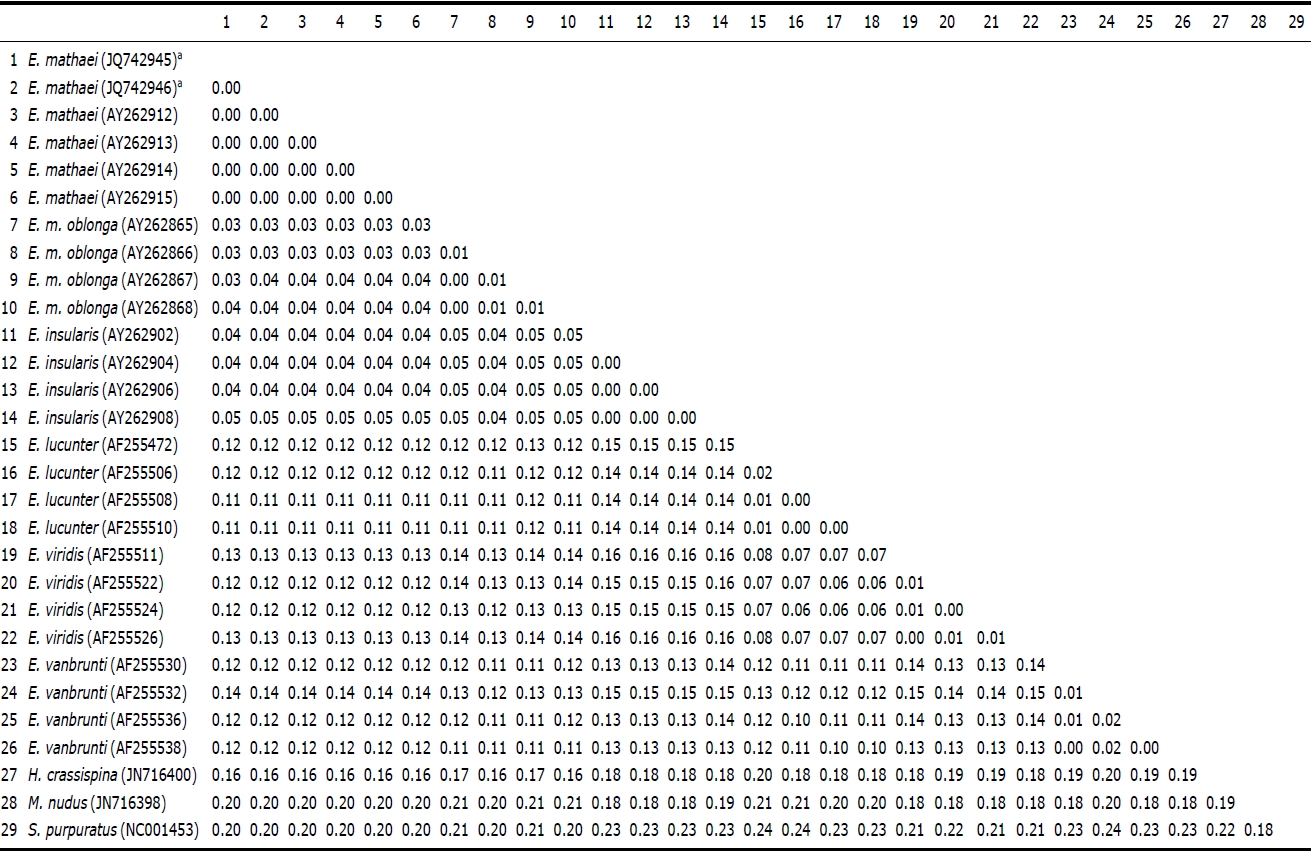
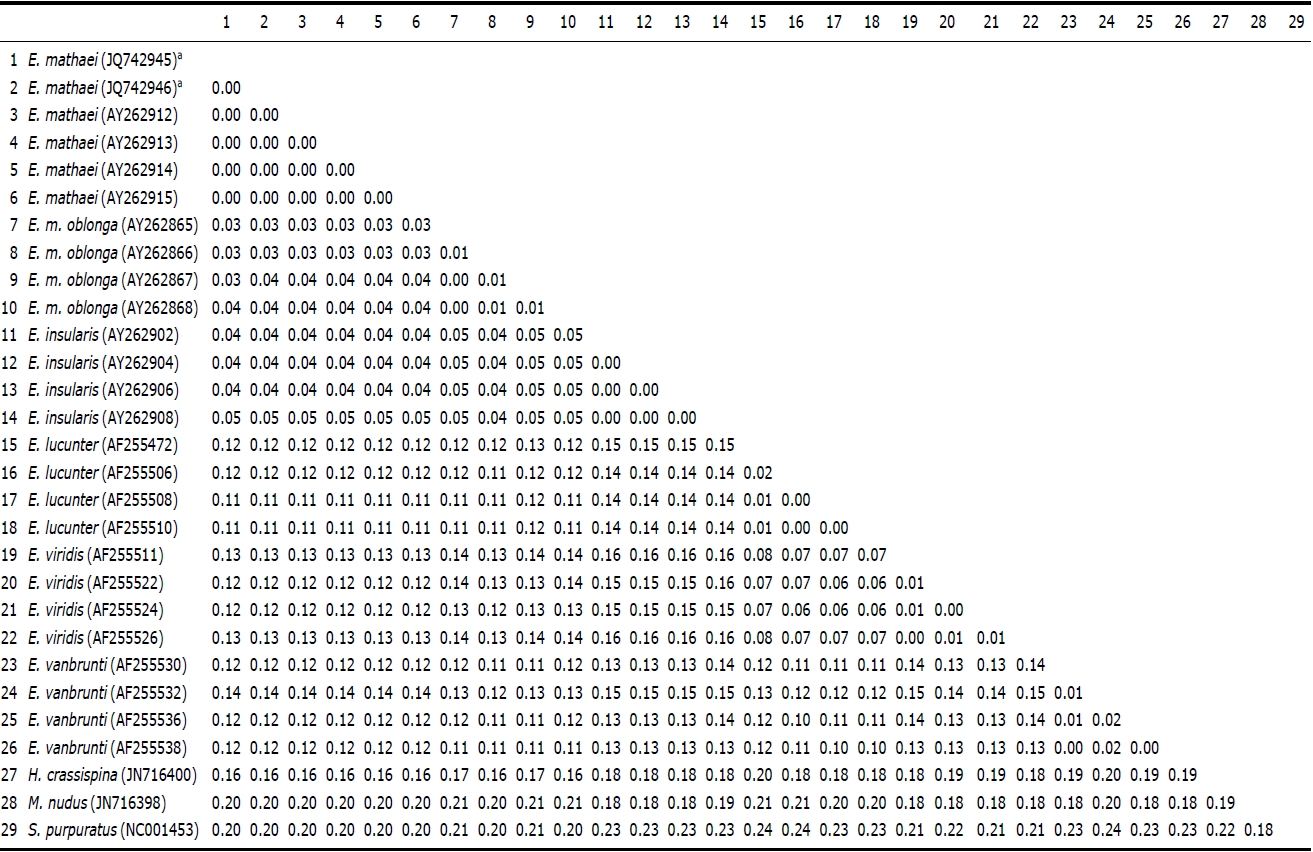
Genetic pairwise distance (p) values were estimated by the Kimura 2-parameter model and were compared (Table 2). As a result, no differences were found between the two Korean specimens. The average value of Korean E. mathaei-GenBank E. mathaei was “0.00”. This value was much lower than the distance values of E. mathaei-E. m. oblonga (p=0.03), E. mathaei-E. insularis (p=0.04), E. mathaei-E. lucunter (p= 0.12), E. mathaei-E. vanbrunti (p=0.12) and E. mathaei-E. viridis (p=0.12).
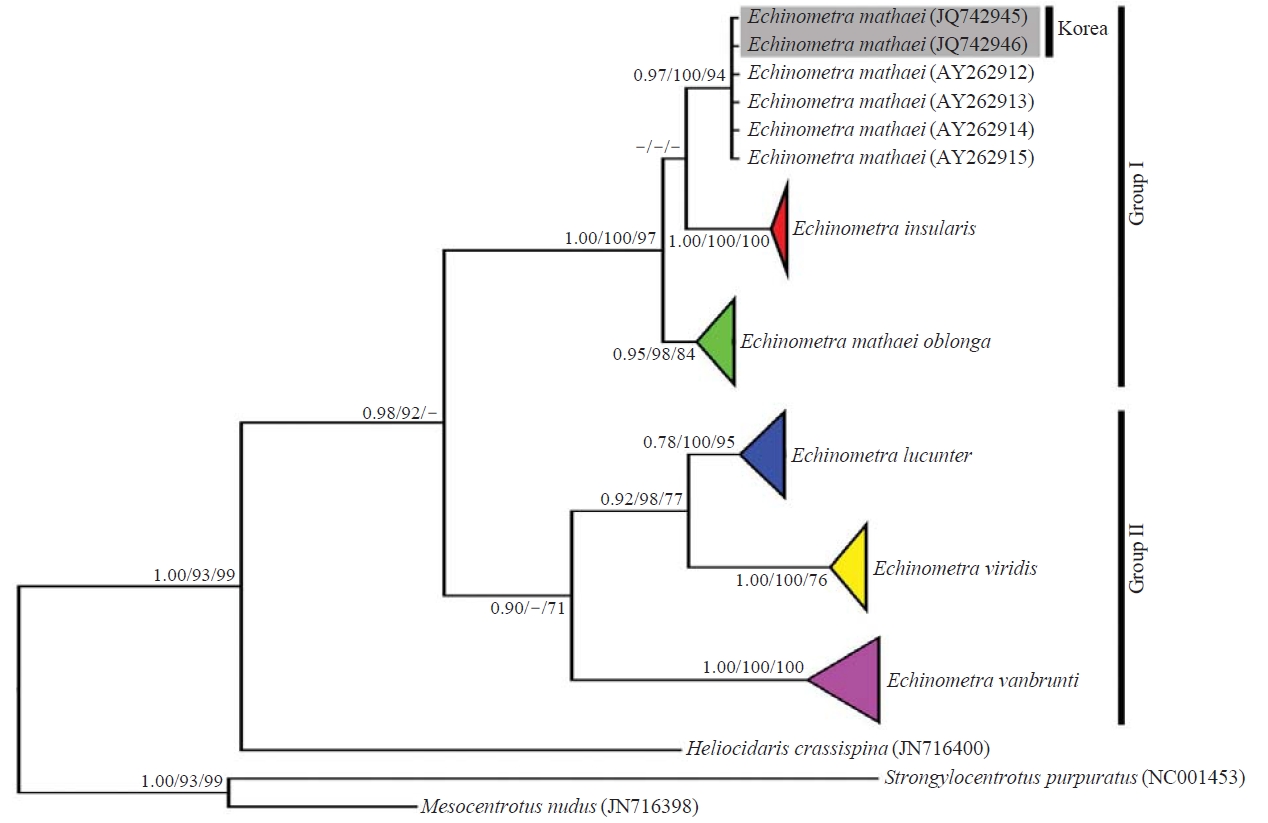
Phylogenetic trees were estimated by BI, NJ and ML methods. Each species of Echinometra established an independent monophyletic clade (Fig. 2). The sequences of Korean E. mathaei formed a distinct monophyletic group with the GenBank E. mathaei, and these sequences were clearly separated from E. insularis and E. m. oblonga. Genus Echinometra was divided into two groups; Group I=(E. m. oblonga (E. mathaei, E. insularis)) and Group II=(E. vanbrubti (E. lucunter, E. viridis)).
Korean name: 1*만두성게속(신칭), 2*만두성게(신칭)
Our specimens were identified as Echinometra mathaei, which is newly recorded in Korea. To date, 13 echinoid species including this species have been reported from Jeju Island, and 32 echinoid species have been recorded in Korea (Shin and Rho, 1996; Shin, 1998; Shin et al., 2006; Lee and Shin, 2011). E. mathaei is represented by great diversity in form of test and spines, as well as in color and form (Mortensen, 1943; Arakaki et al., 1998). Our specimens coincided with these previous morphological descriptions of E. mathaei, but the tubefeet spicules differed slightly from those of Japanese specimens. E. mathaei has C-shaped and triradiate forms of tubefeet spicules (Arakaki et al., 1998; Arakaki and Uehara, 1999), whereas the tubefeet of our specimens contained only C-shaped spicules, but Japanese specimens contained only one C-shaped type or sometimes two tubefeet types (Arakaki et al., 1998). Because of these morphological variations, some studies have investigated the speciation and interspecific relationships of Echinometra species using the mitochondrial COI gene (Palumbi et al., 1997; McCartney et al., 2000; Landry et al., 2003).
The results of the phylogenetic tree (Fig. 2) indicated that the Korean Echinometra specimens clearly established one independent phylogenetic group with those taken from Gen- Bank, and showed no differences in base sequences. These data distinctly support that the Korean Echinometra specimens are E. mathaei. The finding that the genus Echinometra was divided into groups I and II corresponded with the results of previous studies (McCartney et al., 2000; Landry et al., 2003). But, the relationship of group I in the present study was contrary to the results of Landry et al. (2003). The results obtained from 1164 bp of the COI gene dataset analysis using NJ method by Landry et al. (2003) were the same as (E. vanbrunti (E. insularis (E. mathaei, E. m. oblonga))). The results obtained from 930 bp of the COI gene dataset analysis using the NJ method agreed with those of Landry et al. (2003), but the results obtained by the BI and ML methods were the same as those of group I in Fig. 2. Because the results varied according to the analytical methods employed, additional analyses about these relationships are required using other genes.