The Laxmann's shrew (Sorex caecutiens Laxmann 1788), composed of 21 nominal subspecies, is found across the northern part of the Eurasian continent, including the mainland Korean Peninsula, and the peripheral islands of the fareastern Russian Sakhalin and Japanese Hokkaido (Hutterer, 2005). Sorex c. macropygmaeus Miller is distributed in northeastern China and adjacent far-eastern Russia, while S. c. annexus Thomas is found in the central and southern parts of the mainland Korean Peninsula (Jones and Johnson, 1960).
From cytochrome b sequence analyses, Ohdachi et al. (2001) found two distinct subgroups within Sorex caecutiens (one cluster from Hokkaido Island and the other cluster from Eurasian continent and Sakhalin Island), and Ohdachi et al. (2003) reported that the Jeju S. caucutiens formed one of six subclusters within the Continent-Sakhalin-Jeju cluster from the Eurasian continent, the mainland Korean Peninsula, Sakhalin, and Jeju, and the shrews from Jeju were identified as S. caucutiens. Additionally, Ohdachi et al. (2005) classified from morphometric analyses S. caecutiens from Jeju as a new subspecies of S. c. hallamontanus Abe and Oh.
To examine genetic divergences of the two subspecies of Sorex caecutiens from Korea (S. c. hallamontanus in Korean Jeju Island and S. c. annexus from the mainland Korean Peninsula) from cytochrome oxidase I (COI) and cytochrome b sequence analyses, we collected three specimens of S. c. hallamontanus from Jeju Island (one specimen from Kwaneumsa and two specimens from Youngsil) and four specimens of S. c. annexus from the mainland Korean Peninsula (one specimen from Mt. Sobaek and three specimens from Mt. Odae). Small pieces of muscle were collected and preserved in a deep freezer.
Total cellular DNA was extracted using a genomic DNA extraction kit (Intron Co., Seoul, Korea). The COI gene was PCR-amplified using the RonM and NancyM primers, designed by Pfunder et al. (2004), and the PCR thermal cycle employed was as follows: 94℃ for 5 min; 94℃ for 1 min, 53℃ for 1 min, 72℃ for 1 min (35 cycles); and 72℃ for 5 min. The cytochrome b gene was PCR-amplified using the L14724 and H15149 primers, designed by Irwin et al. (1991), and the PCR thermal cycle employed was as follows: 94℃ for 5 min; 94℃ for 1 min, 55℃ for 1 min, 72℃ for 1 min (25 cycles); and 72℃ for 5 min. The amplified products were purified using a DNA PrepMate kit with a silica-based matrix (Intron Co.) The purified PCR products were analyzed with an automated DNA Sequencer (Perkin Elmer 377; Perkin Elmer, Norwalk, CT, USA) at Bioneer Co. (Seoul, Korea).
We first obtained partial COI sequences (429 bp) of the seven specimens from the two subspecies of S. caecutiens in Korea, and these sequences were compared to two haplotypes of S. caecutiens from Maoershan in northeastern China (JF 444466 from Heilongjiang Sheng and JQ599823 from Liaoyeling Sheng), obtained from GenBank. We also obtained complete cytochrome b sequences (1,140 bp) of the seven specimens from the two subspecies of S. caecutiens in Korea, and these sequences were compared to the corresponding 13 sequences of S. caecutiens, obtained from GenBank: two haplotypes (AB028561 and AB028560) from Hokkaido Island, four haplotypes (AB062725 and AB062727 to AB062729) from the mainland Korean Peninsula, two haplotypes (AB0 62731 and AB028554) from Ussuriiskii, Primorye in fareastern Russia, one haplotype (AB028556) from Sakhalin Island, and four haplotypes (AB062721 to AB062724) from Jeju Island.
Sequence alignment, detection of parsimony informative sites, model selection, calculation of nucleotide distances, and tree constructions with 1,000 bootstrapped replications were carried out using MEGA5 (Tamura et al., 2011): the Jukes Cantor (JC) model , which showed the lowest Bayesian information criterion scores, was chosen, and maximum likelihood trees were constructed. In addition, another maximum likelihood trees were constructed by PAUP version 4.0b (Swofford, 2002): HKY85 substitution model was selected by the PAUP, and heuristic searches were performed with step-wise addition options. Sorex isodon (AB062736 for cytochrome b and JF444467 for COI) was used as outgroup.
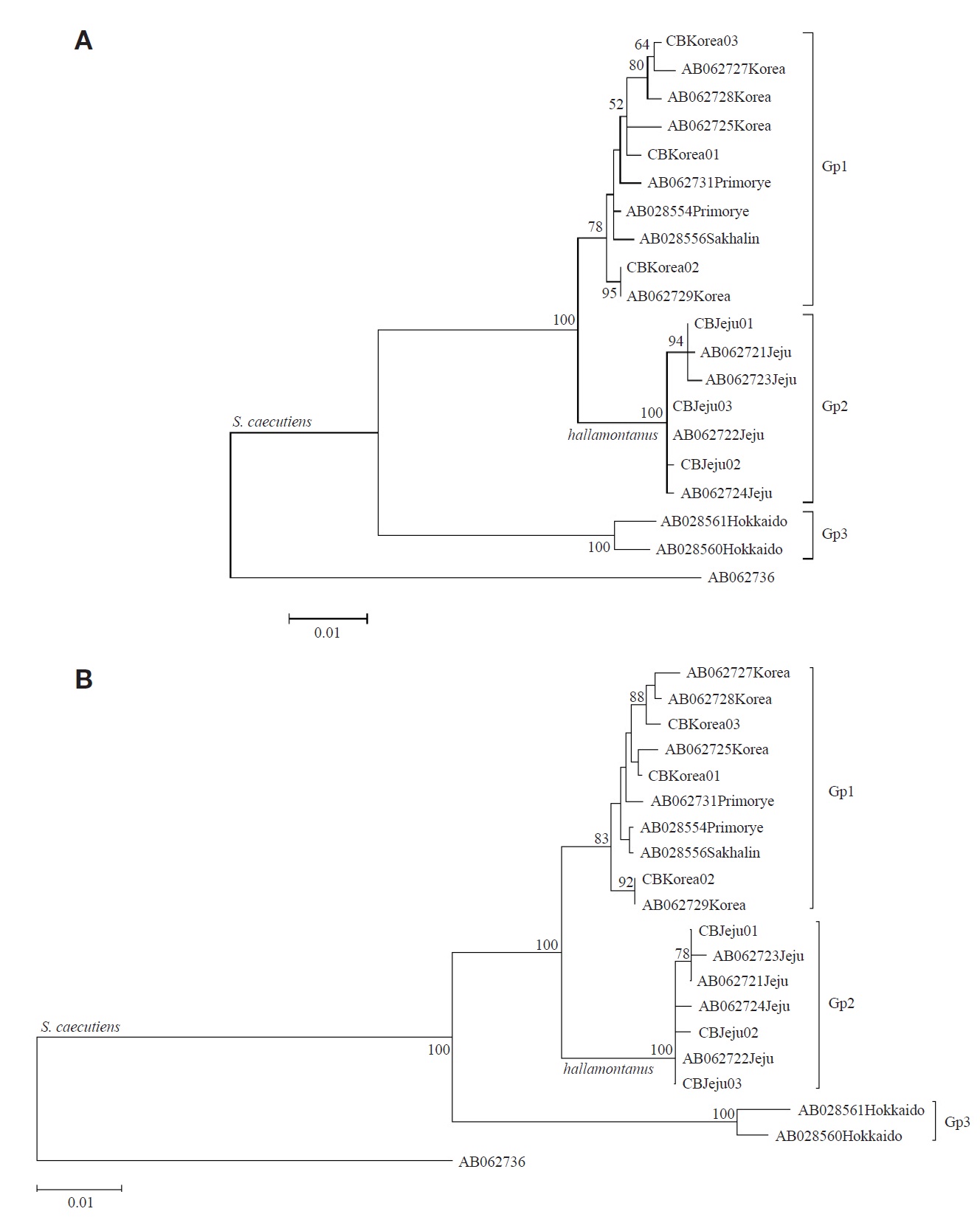
From the seven cytochrome b sequences (1,140 bp), three haplotypes (CBJeju01 to CBJeju03) were identified from S. c. hallamontanus in Jeju Island, and three haplotypes (CB Korea01 to CBKorea03), from S. c. annexus in the mainland Korean Peninsula (six cytochrome b haplotypes of two S. caecutiens subspecies are deposited in GenBank under accession nos. JX192946-JX192951). Within the 19 cytochrome b haplotypes of S. caecutiens (six haplotypes from this study and 13 haplotypes from GenBank), 104 sites (9.12%) were variable, and 82 sites (7.19%) were parsimony informative. Maximum likelihood trees with the 19 haplotypes of S. caecutiens are shown in Fig. 1. All 19 haplotypes were subdivided into three distinct subgroups: ten haplotypes from Primorye, the mainland Korean Peninsula, and Sakhalin (Gp 1, Continent- Sakhalin clade), seven haplotypes from Jeju Island (Gp 2, Jeju clade), and two haplotypes from Hokkaido (Gp 3, Hokkaido clade). The average JC distances between Gps 1 and 2 was 2.07%, between Gps 1 and 3 was 6.27%, and between Gps 2 and 3 was 7.03%. In addition, Gp 2 was distinct from Gp 1, with 11 fixed site differences (site nos., 47, 178, 216, 219, 270, 609, 618, 690, 717, 726, and 898) among 29 parsimony informative sites, and the average JC distance between seven haplotypes from the mainland Korean Peninsula (Gp 1, in part) and other three haplotypes from Primorye and Sakhalin (Gp 1, the rest) was 0.69%.
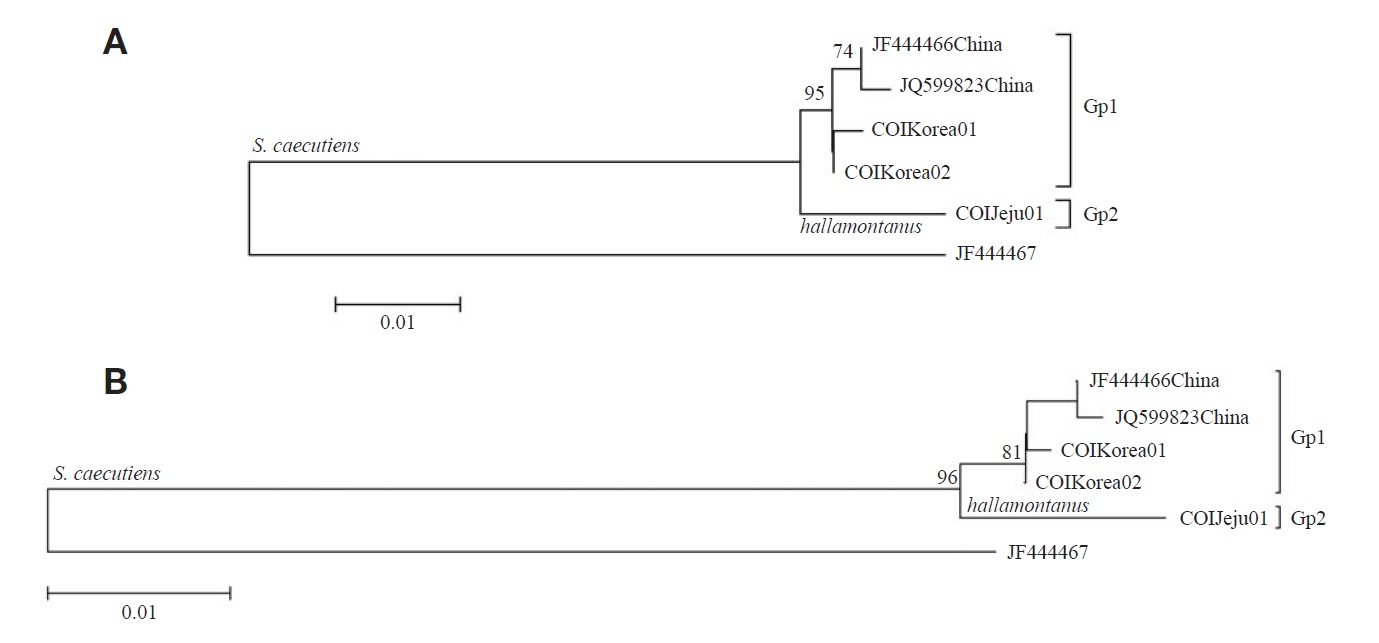
From the seven COI sequences (429 bp), one haplotype (COIJeju01) was identified from S. c. hallamontanus in Jeju Island, and two haplotypes (COIKorea01 and COIKorea02) were, from S. c. annexus in the mainland Korean Peninsula (three COI haplotypes of two S. caecutiens subspecies are deposited in GenBank under accession nos. JX192952-JX1 92954). Maximum likelihood trees with the five haplotypes of S. caecutiens (three haplotypes from this study and two haplotypes from GenBank) are shown in Fig. 2. One monogenic haplotype of S. c. hallamontanus from Jeju Island (Gp 2) was different from four haplotypes of other S. caecutiens from the mainland Korean Peninsula and northeastern China (Gp 1): the average JC distance between two haplotypes from the mainland Korean Peninsula (Gp 1, in part) and two haplotypes from northeastern China (Gp 1, the rest) was 0.47%, but the distance between two subgroups (Gps 1 and 2) was 1.57%.
In this study with the cytochrome b gene sequences (Fig. 1), we found that S. caecutiens is composed of three distinct subgroups (Hokkaido subgroup from Hokkaido Island, Gp 3; Continent-Sakhalin subgroup from Primorye, the mainland Korean Peninsula, and Sakhalin Island, Gp 1; and Jeju subgroup from Jeju Island, Gp 2). The Jeju subgroup (S. c. hallamontanus) was distinct from the other two subgroups (Hokkaido and Continent-Sakhalin), with the average nucleotide distance of 7.03% and 2.07%, respectively. However, Ohda-
chi et al. (2003) reported from their cytochrome b sequence analysis that S. caecutiens is composed of two distinct subgroups (one cluster from Hokkaido Island and the other cluster from Eurasian continent, Sakhalin Island, and Jeju Island), and that four shrews of Sorex caecutiens from Jeju Island formed one of six subclusters within the Continent-Sakhalin- Jeju cluster from Eurasian continent, the mainland Korean Peninsula, Sakhalin, and Jeju.
Pfunder et al. (2004) noted that COI gene can be used for species identification of shrews and voles. From this COI sequence analysis with S. caecutiens (Fig. 2) S. c. hallamontanus from Jeju Island (Gp 2) was distinct from other S. caecutiens in the mainland Korean Peninsula and northeastern China (Gp 1), with the average JC distance of 1.57%. Thus, we concluded that endemic Jeju S. c. hallamontanus is one of three clades within S. caecutiens, with concordant genetic distinctness in mtDNA cytochrome b and COI sequences.
Island populations should diverge over time (genetically and morphologically) from the respective mainland species populations (Johnson et al., 2000), although at the end of the last glacial, large areas of continental shelf were dry land, facilitating exchange of plant and animal species by land bridge connections to what are now isolated islands (Lomolino et al., 2010). Thus, we considered that genetically distinct S. c. hallamontanus from Jeju Island has been geographically isolated even during the last glacial.
A subspecies is an aggregate of phenetically similar populations of a species differing taxonomically from other populations of that species (Mayr and Ashlock, 1991), and it was advocated that a classification should reflect all available characters distributed as widely and evenly as possible over the organisms studied (Huelsenbeck et al., 1996). Ohdachi et al. (2005) classified S. caecutiens from Jeju Island as a new subspecies of S. c. hallamontanus from morphometric analyses, whereas Tatsuo et al. (2005) reported that chromosomal characteristics of S. caecutiens from Jeju Island were essentially identical to those of S. caecutiens from the Eurasian continent. In this paper with cytochrome b and COI sequence analyses (Figs. 1, 2), we found that S. c. hallamontanus is a distinct clade, which is genetically divergent from other populations of S. caecutiens. The importance of examining both nuclear and mitochondrial loci when attempting to elucidate patterns of genetic structure has been previously mentioned (Rubinoff and Holland, 2005), and we propose further systematic analyses with nuclear DNA sequences to confirm our findings.
Bradley and Baker (2001) noted that a genetic distance of < 2% based on the cytochrome b gene was typical of a population and infraspecific variation. Ohdachi et al. (2003) reported from their cytochrome b sequence analysis that phylogenetical relationships among the other five subclusters within the Continent-Sakhalin-Jeju cluster do not always reflect the geographical proximity of their capture locations. In addition, in this study (Figs. 1, 2) S. c. annexus from central and southern part of the mainland Korean Peninsula (Gp 1, in part) was not divergent from S. c. macropygmaeus from northeastern China and nearby Russia (Gp 1, the rest), with the average JC distance of 0.69% in the cytochrome b sequences and the JC distance of 0.47% in the COI sequences. On the other hand, Jones and Johnson (1960) noted that S. c. annexus from the mainland Korean Peninsula is larger in morphological characters than S. c. macropygmaeus from northeastern China and adjacent Russia. Though the mainland Korean S. c. annexus has been considered as an endemic Korean subspecies, our results based on the two mitochondrial gene sequences revealed that the mainland Korean subspecies is not divergent from its neighboring northeastern Chinese subspecies of S. c. macropygmaeus, requiring reexamination for the subspecies status of S. c. annexus.