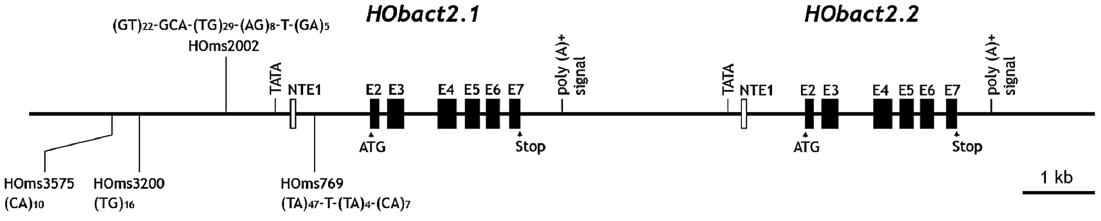
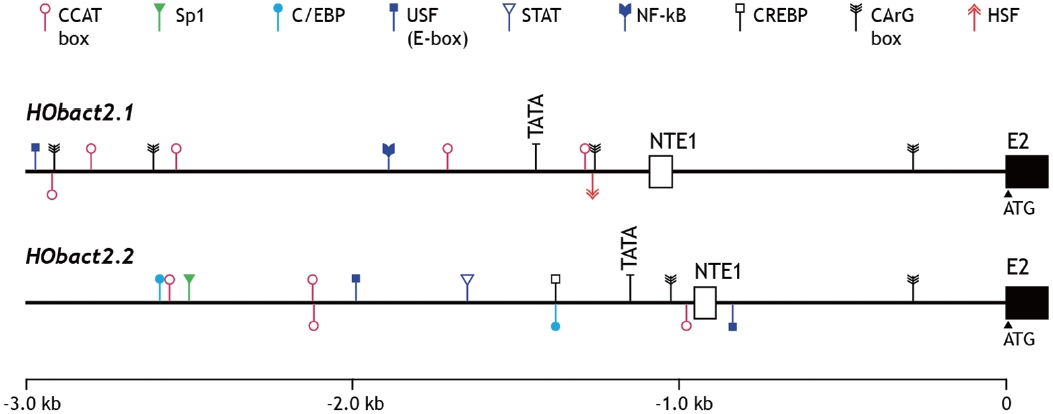
We characterized a cytoplasmic actin gene locus in greenling Hexagrammos otakii (Scorpaeniformes). Genomic clones isolated from the greenling DNA library contained two homologous cytoplasmic actin gene copies (HObact2.1 and HObact2.2) in a tail-to-head orientation. Their gene structure is characterized by six translated exons and one non-translated exon. Exon-intron organiza-tion and the nucleotide sequences of the two actin gene isoforms are very similar. However, only the HObact2.1 isoform contains microsatellite-like, dinucleotide repeats in the 5′-flanking region (named HOms2002) and intron 1 following the non-translated exon 1 (named HOms769). One microsatellite locus (HOms769) was highly polymorphic while the other (HOms2002) was not. Based on bioinformatic analysis, different transcription factor binding motifs are related to stress and immune responses in the two actin isoforms. Semiquantitative and real-time reverse transcription-PCR assays showed that both isoform transcripts were detect-able ubiquitously in all the tissues examined. However, the basal expression levels of each isoform varied across tissues. Overall, the two isoforms showed a similar, but not identical, expression pattern. Our data suggest that the cytoplasmic actin genes may be the result of a recent duplication event in the greenling genome, which has not experienced significant subfunctionalization in their housekeeping roles.
Cytoskeletal actin provides a mechanical support for the cell and mediates cell motility and organelle movement, which are associated with several dynamic cellular activities such as morphological and developmental changes, macromolecule transport, and chemotaxis (Reisler and Egelman, 2007). Fur-thermore, actin is involved in signal transduction (Percipalle and Visa, 2006), RNA localization (Kustermans et al., 2008), apoptosis (Franklin-Tong and Gourlay, 2008), and nitric oxide (NO) regulation (Ji et al., 2007). In addition, the 5′-flanking region of cytoskeletal actin genes is capable of driving the ex-pression of downstream structural genes when it is included in a chimeric transgene construct as a regulatory element (Nam et al., 2008).
In teleosts, genetic determinants of cytoskeletal actin have been isolated from various species belonging to a wide array of taxonomic positions. Although the coding regions of cyto-skeletal actin are well characterized, their genomic organiza-tion and upstream regulatory regions require further investi-gation (see Venkatesh et al., 1996; Cho et al., 2011). Teleosts experienced whole-genome duplication in their evolutionary history and many teleost species may exhibit additional, lo-cus-specific gene duplications in their genomes (Jaillon et al., 2004). Hence, the teleost group likely encodes diverse paralog isoforms in their genomes, and some of these paralogs may undergo subfunctionalization or neofunctionalization. In ver-tebrates, the actin multigene family evolved into six or more isoforms from a single ancestral form (Miwa et al., 1991; Venkatesh et al., 1996), and previous studies have suggest-ed that a greater spectrum of actin isoforms may exist in the fish genome compared to representative mammalian species (Venkatesh et al., 1996). Duplicated (diverged) actin genes are known to exist in the teleost genomes, but their organization has not been extensively studied.
Greenling
>
Fish specimens and nucleic acid libraries
Greenling specimens were purchased from a local fish farm and transferred to a laboratory culture station. Genomic DNA samples were obtained from a fin clip or whole blood using the conventional SDS/proteinase K method followed by etha-nol precipitation. To construct the greenling cDNA library, total RNA was extracted from liver and brain tissues using the RNeasy Midi Kit (Qiagen, Hilden, Germany). After puri-fication of the poly(A)+ RNA fraction using the mRNA Isola-tion Kit (Promega, Madison, WI, USA), an aliquot (5 μg) of poly(A)+ RNA was used as the template for cDNA synthesis. Construction of the cDNA libraries was performed using the Lambda Zap cDNA Synthesis Kit (Stratagene, La Jolla, CA, USA) according to the manufacturer’s instruction. The ge-nomic DNA library was constructed using whole blood DNA purified from a male adult using the Lambda Gem 11 Cloning System (Promega) according to manufacturers’ protocol.
>
Isolation of genomic genes using DNA library screening
Based on our preliminary expressed sequence tag (EST) analysis of greenling brain and liver cDNA libraries (unpub-lished data), an EST clone with significant homology to ver-tebrate cytoplasmic actin was identified and used as a probe to perform a series of filter screenings of the greenling ge-nomic DNA library. The EST clone (0.6 kb) was PCR-labeled with digoxygenin-11-dUTP (Roche Applied Science, Man-heim, Germany). Approximately 5 × 105 phage plaques were screened by plaque hybridization using the DIG DNA Label-ing and Detection Kit (Roche Applied Science). The selected phages were amplified using KW251
Based on genomic sequence analysis, type-specific reverse transcription (RT)-PCR primer pairs (HOact2.1c-1F/1R and HOact2.2c-1F/1R) were designed to amplify the complete open reading frame (ORF) of each isoform. Total RNA (500 ng) from the brain or gill was used as a template for RT-PCR isolation. Oligonucleotide primers and thermal cycling condi-tions used in this study are summarized to Table 1. The am-plification product was sequenced after TA cloning into the pGEM-T Easy Vector (Promega). Based on the ORF-contain-ing sequences, the 5′- and 3′-end untranslated region (UTR) sequences of each isoform were obtained by vectorette PCR, which was performed using template DNA prepared from greenling brain or liver cDNA libraries (excised stocks) and two vector primers (SK and T7). Binding sites are located at either end of the multiple cloning sites in the phagemid vec-tor (pBluescript SK; Stratagene) used for construction of the cDNA libraries. Isoform-specific primers included in the vec-torette PCR as either forward or reverse primers paired with SK or T7 vector primers were HOact2.1-vec1 and HOact2.2-vec1, respectively. Based on the contig assembly of ORF and ends sequences, we determined the full-length cDNA se-quence of each isoform.
>
Bioinformatic sequence analysis
Using NCBI GenBank BLAST, we searched for orthologs of the full-length cDNA sequences. Putative ORFs of each isoform were identified using ORF Finder ( http://www.ncbi.nlm.nih.gov/gorf/gorf.html ). Predicted molecular mass and theoretical pI values were calculated using the ProtParam tool ( http://www.expasy.org/tools/protparam.html ). Multiple sequence alignments were performed using a ClustalW al-gorithm ( http://www.genome.jp/tools/clustalw/ ). Genomic structure and organization were compared using representa-tive orthologs and/or paralogs obtained from the public data-bases, including GenBank and Ensembl ( http://www.ensembl.org/ ). Putative transcription factor (TF)-binding motifs in the 5′-flanking upstream region of the actin isoforms were iden-tified using TFSEARCH ( http://www.cbrc.jp/research/db/TFSEARCH.html ) and Transcription Element Search System (TESS; http://www.cbil.upenn.edu/cgi-bin/tess/tess ). Identifi-cation of repetitive sequences was performed using the Unipro UGENE ( http://ugene.unipro.ru/ ).
>
Typing of simple sequence repeats (SSRs)
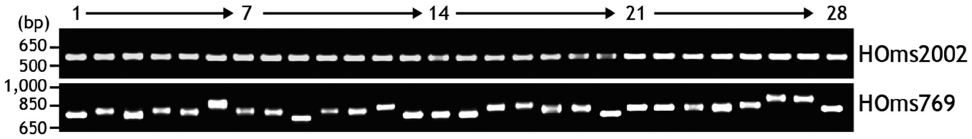
We identified microsatellite-like, SSRs in the noncoding re-gion of one actin isoform. To identify polymorphism among greenling individuals, two PCR primer pairs (HOms769 1F/1R and HOms2002 1F/1R) were designed to amplify each of the two microsatellite regions. In total, 48 individuals ran-domly obtained from three different domestic markets were subjected to microsatellite typing. Genomic DNA was extract-ed from a caudal fin clip (as described above) and 200 ng of purified DNA was used as a template for PCR amplification. The PCR product (5 μL) was separated on 1.5% agarose gel and visualized by ethidium bromide staining to examine any
[Table 1.] Oligonucleotide primers used in this study
Oligonucleotide primers used in this study
polymorphism in PCR product length among individuals.
>
Tissue distribution and basal expression of actin transcripts using a RT-PCR assay
We performed real-time RT-PCR to examine the tissue dis-tribution and basal expression of each actin isoform transcript. Total RNA was extracted from ten somatic tissues including the brain, eye, fin, gill, heart, intestine, kidney, liver, muscle, and spleen, which were obtained from 12 juvenile individu-als (average body weight, 456 ± 52.0 g), as described above. Total RNA (4 μg) was reverse-transcribed (37℃ for 60 min) into cDNA in a reaction volume of 40 μL using the Omniscript Reverse Transcription Kit (Qiagen) according to the manufac-turer’s instructions. A conserved reverse primer for teleost 18S rRNA (Fi18SrRNA-1R primer) was also included in the RT reaction at a 0.1 μM final concentration to prepare the nor-malization standard across tissues. The RT product (cDNA) was diluted fourfold (for actins) or 40-fold (for 18S rRNA) with sterile distilled water, and 2 μL of the diluted cDNA was used as a template for PCR amplification. Based on the pre-liminary optimization of thermal cycling conditions and PCR efficiency (
(Bio-Rad, Hercules, CA, USA) using the 2× iQ SYBR Green Supermix (Bio-Rad) with a reaction volume of 25 μL. Based on the amplification assays (performed in triplicate), the rela-tive transcript levels of each actin isoform across tissues were normalized against the levels of the 18S rRNA control using the following formula: relative expression = [(1 +
>
Gene structure, genomic organization, and cDNA sequences of greenling cytoplasmic actins
Based on the sequence analysis of genomic DNA library clones followed by PCR isolation, the 14,813-bp genomic re-gion containing two cytoplasmic actin genes was read at least twice in both directions. In the genomic region, two copies of the cytoplasmic actin gene existed tandemly in a tail-to-head organization. These two actin copies were named
All actin isoforms have remarkably similar amino acid sequences and polypeptide lengths across diverse metazoan organisms, demonstrating their high evolutionary constraints (Reece et al., 1992). However, compared to mammals, the te-leostean group possesses a wider spectrum of actin isoforms, in which several isoforms show no clear orthology to known mammalian isoforms (Venkatesh et al., 1996; Kim et al., 2008). For example, of the various cytoplasmic actin isoforms identified in teleosts, the major β-cytoplasmic-1 genes are clearly homologous to the mammalian isoforms (
>
Characteristics of the 5’-upstream region
The 5′-flanking upstream regions (3 kb upstream from the translation start codon ATG) of
sive TF-binding motifs in their regulatory regions (Lee et al., 2009; Cho et al., 2011). In addition, our data are partially sup-ported by the fact that mRNA expression of cytoskeletal actin genes is modulated depending on the developmental stage, stress state, or pathological condition of the mammal. Thus, caution is required when using actin gene expression as an in-variant, internal control for gene expression assays (Chapon-nier and Gabbiani, 2004; Filby and Tyler, 2007; Small et al., 2008).
Of the two
>
Tissue distribution and basal expression levels
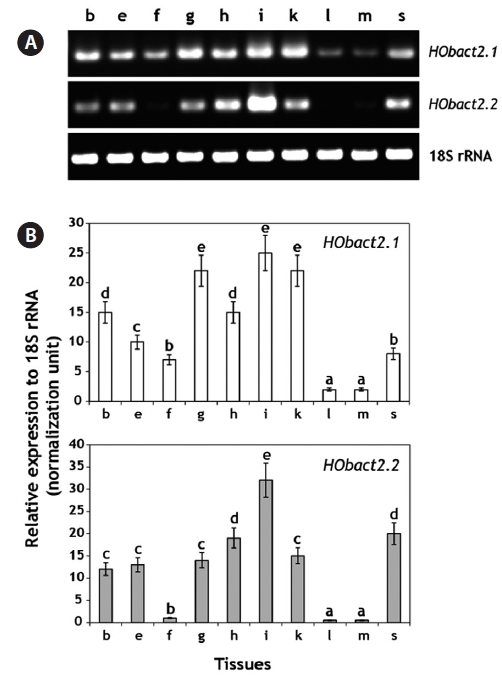
Based on the real-time RT-PCR assay, each greenling actin isoform was detected ubiquitously in all the tissues examined, although the basal expression levels varied among tissues (Fig. 4).
ing the brain, eye, fin, heart, and spleen, showed a moderate level of
In summary, two tandemly existing cytoplasmic actin genes were isolated and characterized from a greenling (