To analyze nucleotide sequence encoding internal transcribed spacer (ITS) regions specific to the Laminariaceae family, genomic DNA was isolated from six brown algae species distributed along the east coast of Korea. These included three species from the Laminariaceae family (Agarum clathratum Dumortier, Costaria costata [C. Agardh] Saunders, and Saccharina japonica Areschoug) and two species from the Alariaceae family (Undaria pinnatifida [Harvey] Suringer and Ecklonia cava Kjellman), both in the order Laminariales, and one species from the family Sargassaceae in the order Fucales (Sargassum serratifolium). Based on a sequence analysis of ITS-1 and ITS-2 for A. clathratum, C. costata, and E. cava, oligonucleotides were designed from the regions that showed sequence conservation in Laminariaceae. Following polymerase chain reaction using three sets of primers, amplification of ITS-1 and ITS-2 was detected in reactions using genomic DNA isolated from the species belonging to Laminariaceae, but not from the species belonging to the other families. The results indicate that this method can be used for the detection and identification of Laminariaceae species.
Algae are photosynthetic organisms found in aquatic environments, and they can be divided into three divisions based on pigment content: Chlorophyta, Pheophyta, and Rhodophyta. The brown algae (Phaeophyta) are a group of mostly marine species found in temperate and colder Northern hemisphere waters. Species belonging to 12 orders in the class Phaeophyceae (Ectocarpales, Ralfsiales, Sphacelariales, Dictyotales, Chordariales, Dictyosiphonales, Scytosiphonales, Cutleriales, Sporochnales, Desmarestiales, Laminariales, and Fucales) are known to be distributed along the coast of Korea (Lee and Kang, 2002). Species belonging to three families (Alariaceae, Chordaceae, and Laminariaceae) in the order Laminariales were found in Korea. The former includes
Commercial production of kelp species has been mostly achieved by mariculture in East Asian countries including China (the largest producer of Laminaria), Korea, and Japan. Korea is the sixth largest macroalgae producer in the world, producing 921,024 MT in 2008, despite the fact that algal species produced in large amounts are mostly cold currentadapted species, meaning that production is restricted during the winter season. For mass production, it is of commercial interest to screen Laminariaceae species in which most of fastgrowing algal species belong.
Genes encoding ribosomal RNAs and internal transcribed
spacer (ITS) regions have been used as molecular markers for species identification and phylogenetic analysis of macroalgae (Bird et al., 1992; Ragan et al., 1994). While a rather conserved 18S rRNA sequence provides information for distinguishing evolutionary distant species, a rapidly evolving ITS sequence has been used for the identification of intraspecific variation. In this study, DNA regions encoding ITS-1 and ITS- 2 of species belonging to Laminariaceae were analyzed to develop a PCR-based species identification method.
Macroalgal species including
Genomic DNA was isolated using previously described nuclei isolation/centyltrimethylammonium bromide (CTAB) methods (Varela-?lvarez et al., 2006). Tissue samples (0.5g) were ground in a mortar in the presence of liquid nitrogen and homogenized in 5 mL STE buffer (400 mM sucrose, 50 mM Tris-Cl pH 7.8, 20 mM ethylenediaminetetraacetic acid (EDTA), 0.2% bovine serum albumin, and 0.2% β-mercaptoethanol). The homogenate was filtered through a 50 μm nylon mesh by squeezing and was subjected to centrifugation at 1,000 g for 20 min. The nuclei pellet was resuspended in 50 μL CTAB buffer (2% CTAB, 2% polyvinylpyrrolidone, 100 mM Tris-Cl pH 8.0, 20 mM EDTA, and 1.4 M NaCl) followed by incubation at 65℃ for 1 h. The suspension was extracted with the same volume of the mixture containing chloroform:isoamylalcohol (24:1) followed by centrifugation at 14,000 rpm for 3 min. DNA in the supernatant was precipitated by the addition of two volumes of ethanol and 0.1 volume of 3 M sodium acetate (pH 5.2) followed by centrifugation.
>
Cloning of the genes encoding ITS-1
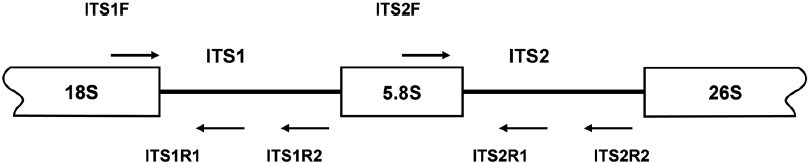
PCR amplifications of the regions corresponding to the inner segments of ITS-1 and ITS-2 were carried out with sets of primers including ITS1F together with the species-specific primers ITS1R1 or ITS1R2, and ITS2F together with ITS2R1 or ITS2R2, respectively. Amplification of ITS-1 was carried out in a 20 μL PCR mixture including a HiQ-PCR Mix, genomic DNA (0.1 μg), and 1 μM primers. The reaction was carried out with an initial denaturation at 95℃ for 5 min followed by 30 cycles consisting of denaturation at 94℃ for 30 s, annealing at 60℃ for 30 s, and extension at 72℃ for 30 s, together with a final extension at 72℃ for 5 min. PCR amplification using ITS2F and ITS2R1 primers was also carried out under the same condition except an annealing temperature at 65℃. Amplified PCR products, resolved upon agarose gel electrophoresis, were purified by gel extraction, cloned into the pTOP TA V2 cloning vector, and then transformed into
The nucleotide sequences obtained were aligned with other ITS sequences using ClustalW (Thompson et al., 1994). The
accession numbers of ITS-1and ITS-2 sequences, acquired from the NCBI GenBank database (Guiry and Guiry, 2011) were as follows:
The production of macroalgae has been increasing in recent years as its demand for commercial application has increased.
In Korea, the production of major algal species including
Algal species are classified by phenotypic characteristics as well as genetic markers (Yoon et al. 2001; Saunders, 2005). Differences in the morphological characteristics including the presence of mucilaginous organs in the sporophyte, lack of an eyespot in meiospores, and uniflagellation of the sperm exist among the families in the order Laminariales (Kawai and Sasaki, 2000). The Laminariaceae family is characterized by the presence of a distinct stipe, paraphyses without hyaline appendages, and the absence of outgrowths or splitting of the transition zone. In particular,
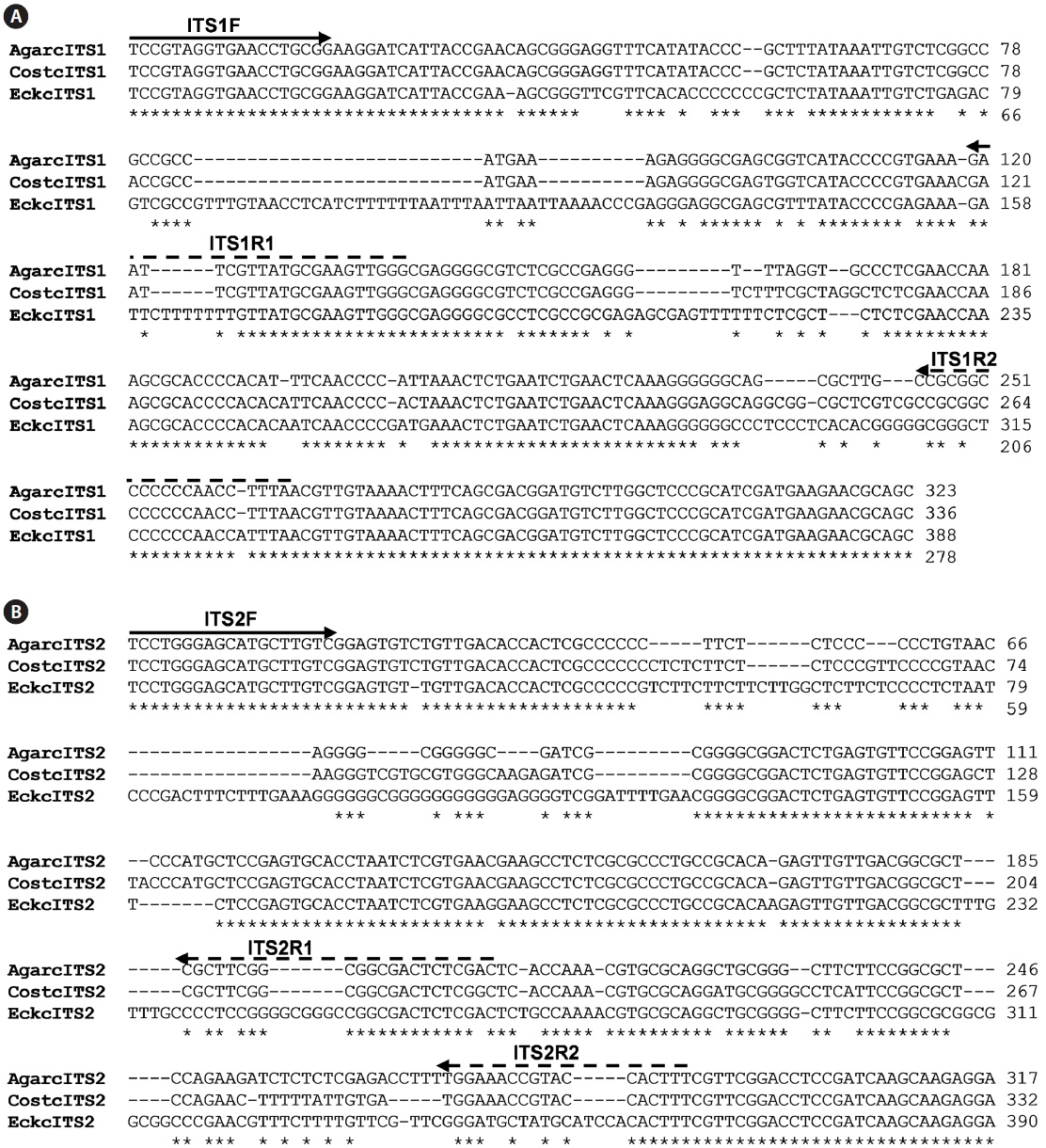
The isolation of genomic DNA from macroalgae and its subsequent use in PCR are known to be hindered by polysaccharides and phenolic compounds (Koonjul et al., 1999; Varma et al., 2007). Among the methods used for genomic DNA isolation from macroalgae (Varela-?lvarez et al., 2006; Hoarau et al., 2007; Snirc et al., 2010), the STE and CTAB buffer method was used for the species tested in this study. Upon confirming the integrity of genomic DNA by agarose gel electrophoresis (data not shown), DNA was used as a template for amplification of the regions corresponding to ITS. The typical ribosome in algae is composed of four rRNAs of which the rRNA cluster of 18S-5.8S-26S is transcribed from a single transcriptional unit (Coleman and Mai, 1997; Yotsukura et al., 1999; Torres-Machorro et al, 2010). In this study, PCR amplification of regions encoding ITS-1 and ITS-2 using a genomic DNA template and primers (ITS1F/ITS1R and ITS2F/ ITS2R) resulted in 250-300 bp ITS-1 and ITS-2 fragments depending on the species, similar to the sizes reported from other macroalgal species (data not shown). At least three independent clones were analyzed to confirm the ITS sequences presented as many copies in the genome. A sequence comparison of ITS-1 and ITS-2 was first performed with three closely related species,
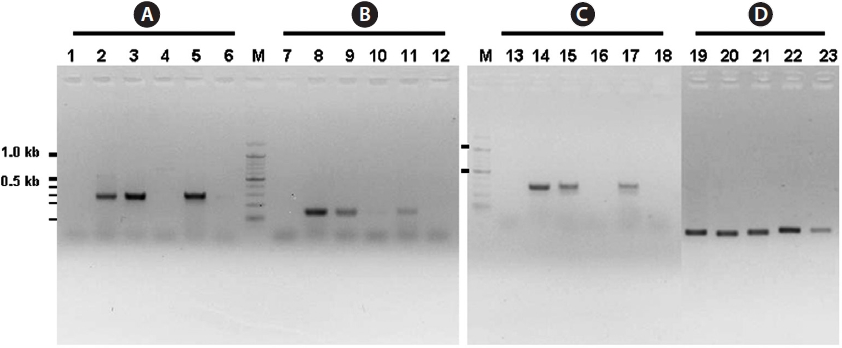
The inner segments of ITS-1 and ITS-2 were amplified using primers specific to the Laminariaceae family designed in this study. In PCRs using three sets of primers, amplified products were detected in reactions using genomic DNA isolated from
The present study clearly showed that only genomic DNA isolated from the family Laminariaceae could be amplified by PCR, indicating that PCR using the primer sets of ITS1F together with ITS1R1 or ITS1R2, and ITS2F together with ITS2R1, can be used for the specific detection of Laminariaceae. This principle can be applied to develop other familyspecific detection methods.