The marine unicellular red algal genus Rhodella was established in 1970 by L. V. Evans with a single species R. macu-lata based on nuclear projections into the pyrenoid. Porphyridium violaceum was described by P. Kornmann in 1965 and transferred to Rhodella by W. Wehrmeyer in 1971 based on plastid features and the non-parietal position of the nucleus. Molecular and fine structural evidences have now revealed that Rhodella maculata and R. violacea are one species, so R. violacea has nomenclatural priority and the correct name is Rhodella violacea (Kornmann) Wehrmeyer. The status of families within Rhodellophyceae was examined. The order Dixoniellales and family Dixoniellaceae are emended to include only Dixoniella and Neorhodella. The order Rhodellales and family Rhodellaceae are emended to include Rho-della and Corynoplastis. Glaucosphaera vacuolata Korshikov and the Glaucosphaeraceae Skuja (1954) with an emended description are transferred to the Glaucosphaerales ord. nov.
The genus
In the 1980s, two more unicellular red algae were iso-lated, characterized by TEM and assigned to the genus
Yoon et al. (2006) established six classes of red algae in subphylum Rhodophytina, three of which include uni-cellular forms, the Porphyridiophyceae (
This current work presents molecular, confocal micros-copy, and electron microscopy details on culture strains of a unicellular red alga identified as
Procedures for isolation and culture were as described in West (2005). Isolate JAW 2347 (CCMP 3129) was ob-tained from a mud sample in a
>
Confocal and Nomarski microscopy
Fixation, staining, and observation methods were par-tially described in Yang et al. (2011). Cultured cells were transferred to 1.5 mL microtubes with 0.5 mL of culture medium, fixed with an equal volume of 3% paraformal-dehyde in PHEM buffer [60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2(6H2O), pH 7.4] for 15 min. Ten μL of fixed cells were placed on a glass slide coated with 0.1 w/v% poly-L-lysine and incubated for 10 min at room temperature. Then equal volumes of SlowFade Antifade (Invitrogen, Carlsbad, CA, USA) and 1,000× diluted SYBR Green I (Invitrogen) were added and incubated for 15 min. Confocal images were taken with an inverted microscope Axiovert 100M with LSM 510 laser scanning equipment (Carl Zeiss AG, Jena, Germany) at the University of Tsuku-ba, and an Axio Observer LSM 700 instrument at Bigelow Laboratory for Ocean Sciences. Plastid autofluorescence and SYBR Green I fluorescence were detected with a 585 nm long pass filter and a 505 to 530 nm band pass filter, respectively, in the excitation line of a 488 nm argon laser and 543 nm He / Ne laser using a single-track mode. Liv-ing cells were observed with a Nikon microscope (Eclipse E600) equipped with Nomarski interference optics (Nikon Co. Ltd., Tokyo, Japan).
>
Transmission electron microscopy
Cells from cultures (grown in window light in 15 psu von Stosch enriched seawater) were filtered onto poly-L-lysine coated 0.45 μm Millipore filters (Bedford, MA, USA) and fixed for 2 h at ambient temperature in 2% glutaraldehyde in a 0.1 M phosphate buffer solution (pH 6.8) with 0.15 M sucrose. Following buffer rinses samples were post-fixed 2 h in the same buffer in 1% OsO4 at 4℃, rinsed thoroughly in H2O, left in 50% acetone for 30 min, and stored in a 70% acetone-2% uranyl acetate solution at ambient temperature for 22 h. Samples were then further dehydrated in a graded acetone series, infiltrated gradu-ally, embedded in Embed 812 resin (Electron Microscopy Sciences, Hatfield, PA, USA), and polymerized at 70℃ for 2-3 days. Thin sections were cut with an RMC MT6000-XL ultramicrotome (RMC Inc., Tucson, AZ, USA). Sections were stained for 1 min with lead acetate. A Zeiss EM 109 electron microscope was used for observation and pho-tography.
>
DNA extraction, amplification and sequencing
Genomic DNA was extracted from each culture strain using a DNeasy Plant Mini Kit (Qiagen, Hilden, Ger-many) according to the manufacturers’ instructions. Polymerase chain reaction (PCR) and sequencing were performed with published and newly designed spe-cific primer sets for each gene; psaA130F-psaA1110R, psaA870F-psaA1600R and psaA971F-psaA1760R for
Published sequences were obtained from GenBank and aligned with newly determined sequences using ClustalW implemented SeaView version 4.2.5 (Gouy et al. 2010) and manually refined using Se-Al version 2.0a11 (http://tree.bio.ed.ac.uk/software/seal/). We excluded insertions of the nuclear small subunit rRNA (SSU rRNA) (
The evolutionary model for individual genes was cho-sen using the weighted Akaike information criterion (AIC) implemented in ModelGenerator version 0.85 (Keane et al. 2006). The selected best fitting models were the gen-eral time reversible (GTR) substitution with the gamma distributed rate heterogeneity (G) for DNA data; the LG substitution (Le and Gascuel 2008) with empirical amino acid frequencies (F) and G (LG + F + G model) for protein data. We used an independent model for each partition of the concatenated data. For example, it was used GTR + G model for SSU part and separate LG + F + G for
Maximum likelihood (ML) analyses were performed using RAxML version 7.2.8 (Stamatakis 2006). Tree like-lihoods were estimated using 200 independent replica-tions, each with a random starting point. The separate site-specific model was used for partitioned data and the automatically optimized SPR branch rearrangements were used during the rapid hill climbing tree search for each replication. Bootstrap analyses (MLB) were con-ducted using 1,000 replications with the same evolu-
[Table 1.] Taxa list used in present study
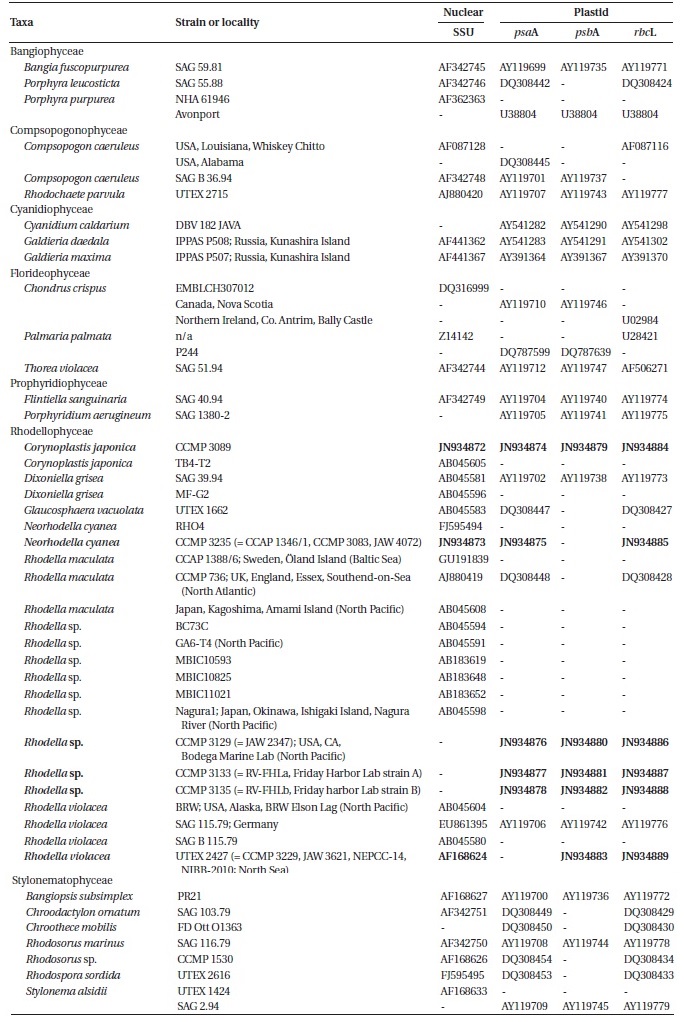
Taxa list used in present study
tionary model setting as that used for the best topology search.
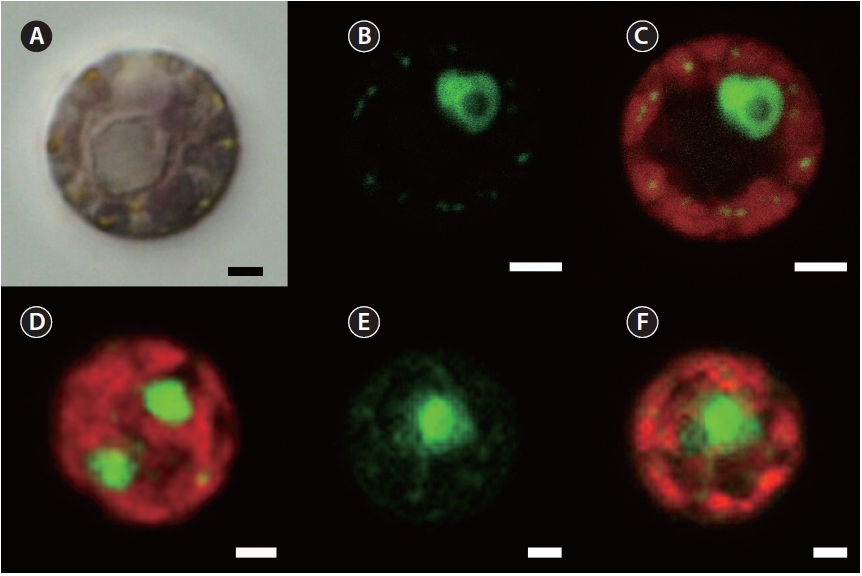
The following observations were made with the culture isolate JAW 2347 (CCMP 3129). SYBR Green stain gave a green fluorescence to nuclear DNA as well as chloro-plast and mitochondria genophores, whereas the whole chloroplast showed a red autofluorescence (Fig. 1). The nucleus of the interphase stage was always eccentric and associated with a pyrenoid (Fig. 1A-C). The single chlo-roplast had its pyrenoid centrally located and had many lobes positioned at the cell periphery. Plastid genophores were spherical to subspherical (about 0.25 μm diameter) and present in the peripheral lobes. Two nuclei were found in some dividing cells (Fig. 1D). All observed char-acters were also found in
>
Transmission electron microscopy
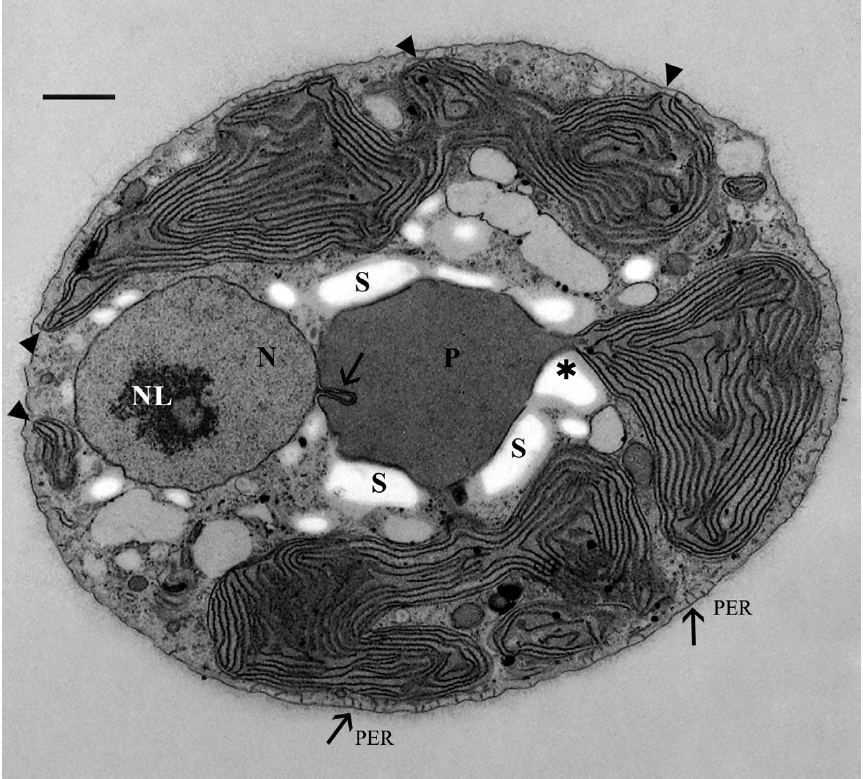
The cells were 8-12 μm in diameter and were coated by thin fibrillar material (Fig. 2). A discontinuous layer of ER was present beneath the cell membrane and appeared interconnected with the cell membrane by short tubules (Fig. 2 & 3D), as seen by Patrone et al. (1991) in
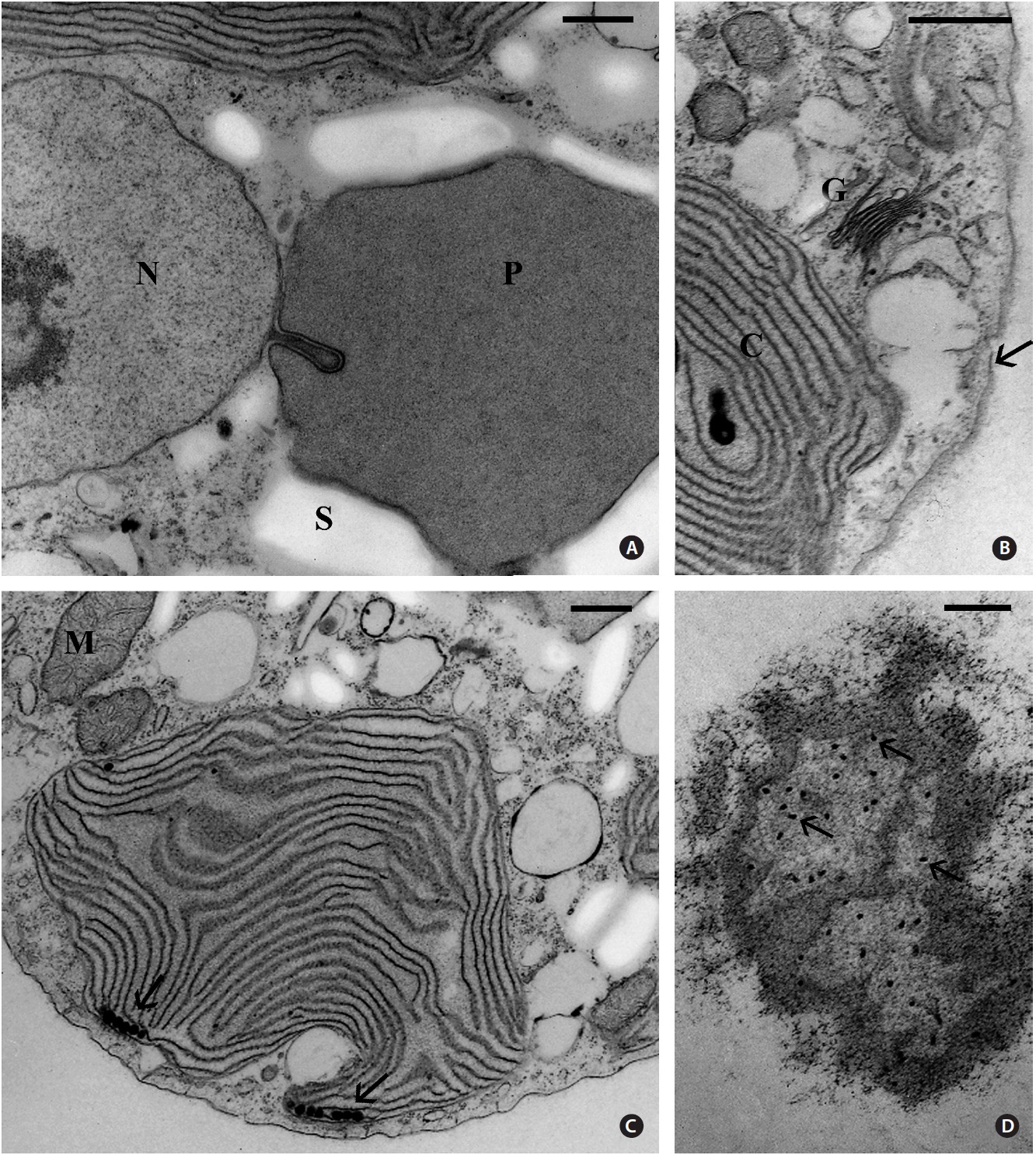
Usually one pyrenoid was found in each cell but a few cells contained two pyrenoids. The pyrenoids were com-monly larger than nuclei and were often bordered by starch that was also seen elsewhere in the cell, frequently near nuclei (Fig. 2). The moderately electron dense py-renoid matrix was devoid of thylakoids (Fig. 2 & 3A). As observed in two-dimensional sections, 1-4 chloroplast lobes extended from the pyrenoid throughout the cy-toplasm. Thylakoids were abundant and maintained consistent spacing from each other (Fig. 2, 3B & C), but phycobilisomes were not usually clearly seen. Peripheral thylakoids were absent (Fig. 2 & 3C). Electron dense clus-ters of spherical globules (plastoglobuli) were commonly seen at the outermost periphery of chloroplast lobes (Fig. 3C). Profiles of mitochondria were found throughout the cell (Fig. 2, 3B & C) as well as small electron transpar-ent vacuoles (Fig. 2 & 3B). Golgi bodies were somewhat sparse and usually located at peripheral cell regions. All observed Golgi bodies had fused or closely appressed cis-ternae (Fig. 3B).
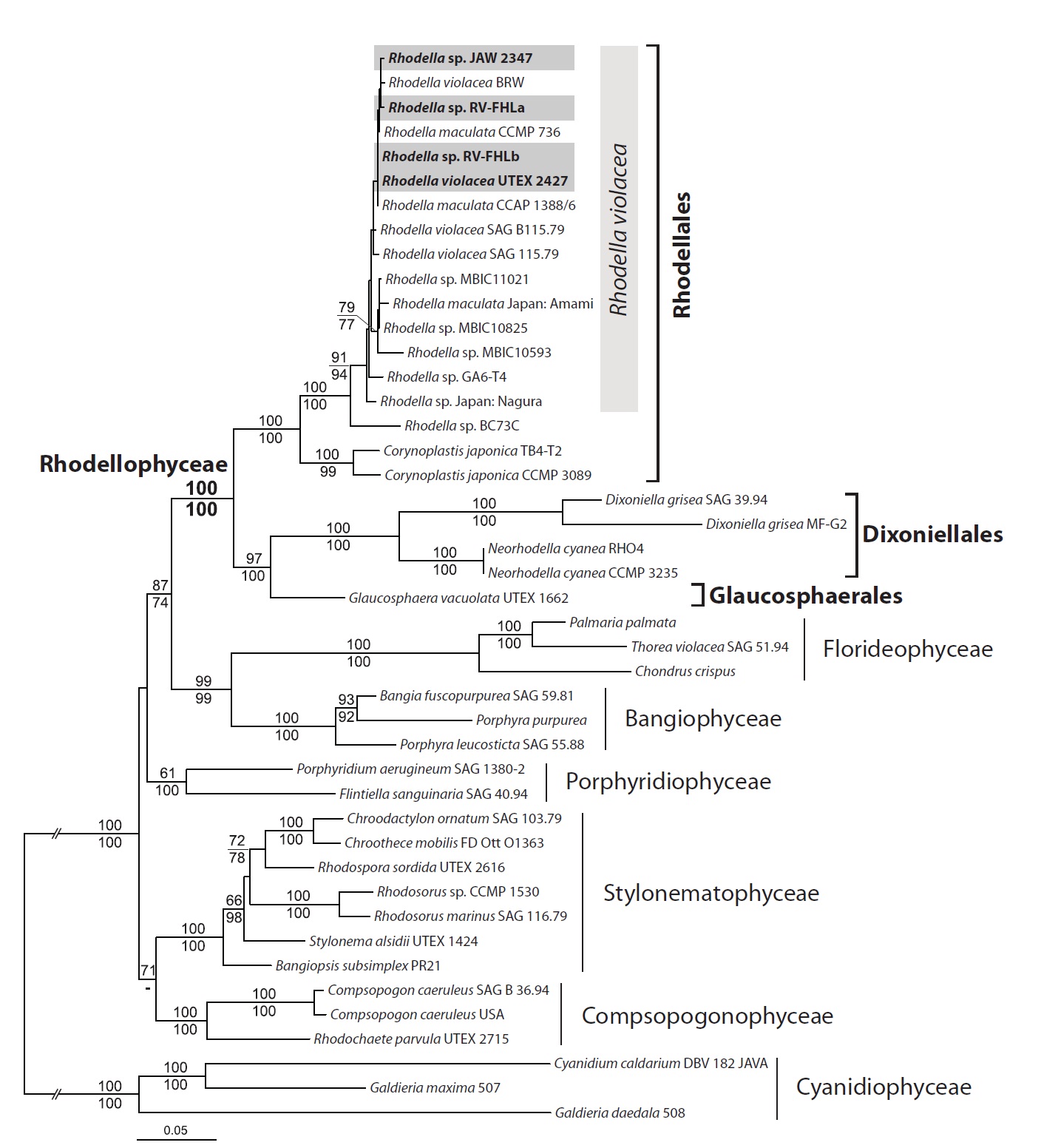
Newly determined rhodellophycean nuclear SSU rRNA and three plastid protein-coding
DNA data were consistent with each other, except for the position of Porphyridiophyceae (Fig. 4). The Porphyrid-iophyceae grouped with a Bangiophyceae / Florideophy-ceae / Rhodellophyceae clade in the DNA + protein mixed data, whereas it grouped together with the Compsopogo-nophyceae / Stylonematophyceae clade in the DNA-only tree. However, this incongruence was not supported sta-tistically (less than 50% bootstrap value in each phylog-eny).
Multigene trees supported the monophyly of Rhodel-lophyceae strongly (MLB and mixed and DNA 100%). Within the rhodellophycean monophyly, two subclades were identified: i) the Rhodellales clade (MLB mixed and DNA 100%) including
Sequence divergences within
Our ultrastructural examination of JAW 2347 revealed two cell characters found in no other unicellular red al-gae except
The only other member of the Rhodellophyceae pos-sessing Golgi bodies associated with ER is
The presence or absence of a peripheral thylakoid is variable in the Rhodellophyceae.
It is obvious that the presence or absence of starch grains closely bordering the naked pyrenoids of
in his study, it is possible that starch could likewise be ab-sent in the protrusion zone of the isolate of
The nuclear protrusions in strain JAW 2347 (CCMP 3129),
The low molecular weight carbohydrate (LMWC) man-nitol is present in all Rhodellophycae including JAW 2347 (CCMP 3129) (Karsten et al. 1999, 2003, Scott et al. 2008, Nitschke et al. 2010).
JAW 2347 (CCMP 3129) is readily recognized as a species of
The concatenated phylogeny (nuclear SSU rRNA + plastid
In the
>
Redefinition of the Rhodellophyceae orders and families
Wynne and Schneider (2010) indicated that the family Dixoniellaceae established by Yokoyama et al. (2009) was invalid because the family Glaucosphaeraceae, which originally included only
Glaucosphaerales ord. nov., Yang, Scott, Yoon and West. The order, family and genus are defined as a unicel-lular freshwater red alga, containing a blue green chloro-plast with a peripheral thylakoid, plastoglobuli clusters, no pyrenoid, active perinuclear Golgi bodies continuous-ly producing and rapidly ejecting small vesicles from the cell, cells larger than most unicellular reds (to 25 μm di-ameter) (Broadwater et al. 1995, Pickett-Heaps et al. 2001, Wilson et al. 2006), LMWCs unknown.
Glaucosphaeraceae Skuja (1954). Same characters as the order. The only taxon at present is
Dixoniellales and Dixoniellaceae Yokoyama et al. (2009). Emended ordinal and family description: single chloroplast with a single or multiple pyrenoids, plasto-globuli at chloroplast periphery, Golgi bodies perinuclear, mannitol as the only LMWC.
Rhodellales Yoon et al. (2006). As originally defined by Yoon et al. (2006) this order includes
Emended ordinal description: unicellular red algae, a single highly lobed plastid with single or multiple pyre-noids, plastoglobuli at plastid periphery, scattered Golgi associated with ER, contain mannitol as a LMWC.
Rhodellaceae Yoon et al. (2006) now contains only two genera