Arthrospira is a commercially important filamentous cyanobacterium with an annual production estimated at over 3,000 tons per year, the largest among microalgae (Pulz and Gross 2004). Found in tropical and subtropical regions in warm lakes with a high carbonate and bicarbonate content, and high pH and salinity (Tomaselli 1997), Arthrospira is a rich source of proteins, minerals, vitamin B12, β-carotene, and essential fatty acids, such as γ-linolenic acid. Owing to its high protein content of up to approximately 60-70% on a dry weight basis, the amino acids present in Arthrospira match the proportions recommended by the Food and Agriculture Organization (FAO) (Dillon et al. 1995). Plus, it does not require any chemical or physical processing in order to become digestible due to the absence of a cellulose cell wall (Vonshak 1997b). Indeed, it can be used as feed for fish, poultry, and farm animals (Belay 1997, Vonshak 1997a). Originating from the taxonomic studies of cyanobacteria by Geitler (1932), some confusion has arisen between the genera Spirulina and Arthrospira (Desikachary and Jeeji-Bai 1996). While the 16S rRNA gene sequences of these two genera indicate that they are only distantly related (Nelissen et al. 1994, Scheldeman et al. 1999), putative species of Arthrospira have been recognized in the so-called ‘Spirulina’ commercial industrial field.
Microscopy identification of cyanobacteria is rapid and sensitive. However, even with skilled and experienced operators, identification is sometimes uncertain. Thus, molecular approaches are particularly useful in the detection and identification of specific strains, especially those that are morphologically identical at the species level. Plus, genetic identification has been used to discriminate nuisance species in cyanobacterial genera, including Microcystis, Anabaena, Nodularia, and Cylindrospermopsis (Wilmotte and Golubic 1991, Neilan et al. 1995).
Phylogenetic analyses based on the 16S rRNA gene and cpcBA-intergenic spacer (cpcBA-IGS) take considerable time and effort, yet they provide certainty in the identification of genera using both the length and the sequence of the cpcBA-IGS. In addition to 16S rRNA gene sequence analyses (Otsuka et al. 1998, Honda et al. 1999, Turner et al. 1999), the genes encoding the major light-harvesting accessory pigment proteins, particularly the phycocyanin operon (cpc), including the IGS between cpcB and cpcA and the corresponding flanking regions (cpcBA-IGS), have also been targeted for phylogenetic studies of cyanobacteria (Neilan et al. 1995, Barker et al. 1999). Furthermore, to determine the phyletic relationships between Arthrospira strains, a region of the phycocyanin locus, namely cpcBA-IGS, has been sequenced, as its nucleotide substitution rate is potentially higher than
that of the widely used 16S rRNA gene sequence (Lyra et al. 2001).
Accordingly, the aim of this study is to clarify the diversity of cultivated strains of Arthrospira using sequence data from a highly variable DNA fragment, including a comparison of the phylogeny of Arthrospira strains based on the 16S rRNA gene and cpcBA-IGS. Plus, to define and delimit the genus Arthrospira, this study also includes other cyanobacteria. Finally, this study attempts to identify a specific molecular marker to fingerprint different strains of Arthrospira and develop tools for species and strain identification.
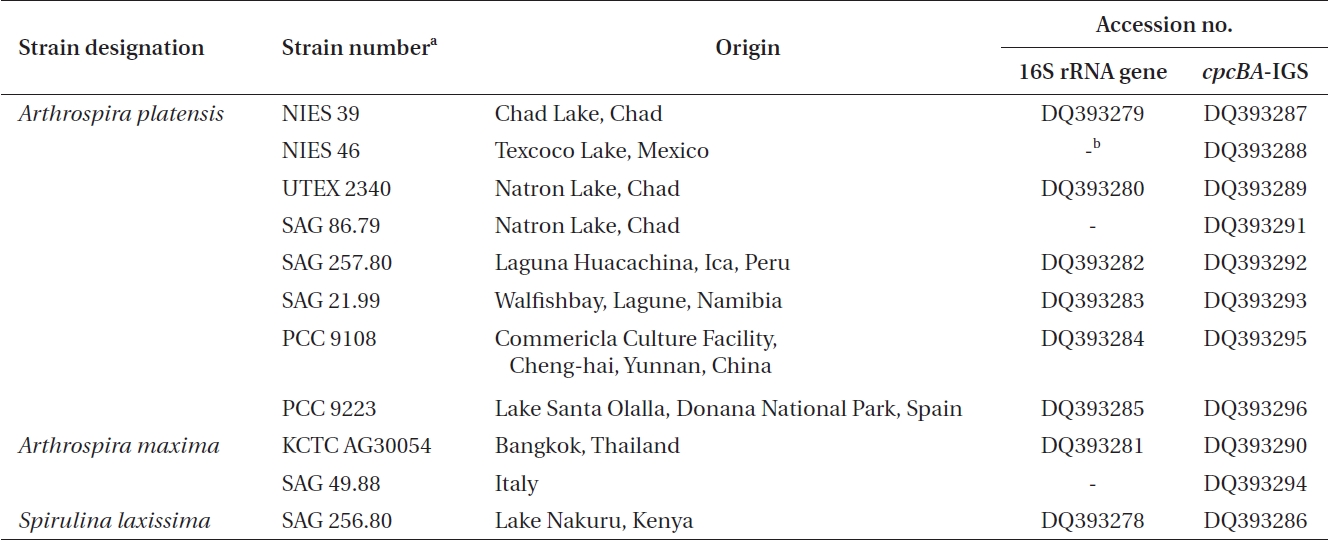
Ten Arthrospira cultures and one Spirulina culture were obtained from different culture collections across the world. Table 1 shows a list of the 10 Arthrospira strains, along with their origin, culture history, and the corresponding DNA database accession numbers for the 16S rRNA gene and cpcBA-IGS. The cultures were maintained in an SOT medium (Choi et al. 2003), containing per liter 16.8g of NaHCO3, 0.5g of K2HPO4, 2.5g of NaNO3, 1g of K2SO4, 1g of NaCl, 0.2g of MgSO4·7H2O, 0.04 g of CaCl2·2H2O, 0.01g of FeSO4·7H2O, 0.08g of Na2EDTA, 0.03 mg of H3BO3, 0.025 mg of MnSO4·7H2O, 0.002 mg of ZnSO4·7H2O, 0.0079 mg of CuSO4·5H2O, and 0.0021 mg of Na2MoO4·2H2O. Stock cultures of 10 mL were kept under low light (20 μmol photons m-2 s-1) and at a constant temperature of 25 ± 1℃.
The cultured strains were harvested using centrifugation, and the genomic DNA extracted following the method of Neilan et al. (1995) using lysozyme and proteinase K. The 16S rRNA gene was then amplified using the universal primers 27F (5′-AGAGTTTGATCMTGCTGGCTCAG-3′) and 1512R (5′-GGYTACCTTGTTACGACTT-3′) (Cho and Giovannoni 2003). The PCR products were purified using a Qiagen QIAquick PCR purification column and cloned using a pDrive vector (Qiagen, Hilden, Germany). The cpcBA-IGS was amplified using primers located within the coding region of cpcB and cpcA, respectively, where the forward primer was CPC1F (5′-GGC KGC YTG YYT GCG YGA CAT GGA-3′) and the reverse primer was CPC1R (5.-AAR CGN CCT TGR GWA TCD GC-3′) (Kim et al. 2006). The amplified fragments (annealing temperature 55℃) were purified using a Qiagen QIAquick PCR purification column and cloned using a pDrive vector (Qiagen). The PCR products were then sequenced using the SP6 and T7 primers with the chain-termination method on an ABI377 automated sequencer (Solgent Ltd., Daejeon, Korea). The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers DQ393278-DQ393296.
The 16S rRNA gene and cpcBA-IGS sequences obtained from the Arthrospira strains were initially compared with sequences available in the National Center for Biotechnology Information database using BLAST network services (http://www.ncbi.nlm.nih.gov/BLAST) to determine their approximate phylogenetic affiliations (Altschul et al. 1997). The sequences were aligned using PHYDIC 3.0, and unambiguously aligned nucleotide positions then used for phylogenetic analyses using PHYDIC 3.0 (Chun and Goodfellow 1995). The similarity values between the sequences were calculated from distance matrices by reversing the Jukes-Cantor distance formula (Jukes and Cantor 1969). Phylogenetic trees were then inferred by neighbour joining (NJ) (Saitou and Nei 1987) using the Kimura two-parameter model. The resulting NJ tree was evaluated by bootstrap analyses based on 1,000 resamplings. Due to the spacer variability, a phylogenetic analysis of the matrix was also performed using just the two coding regions. Finally, an overview of the phylogenetic position of Arthrospira in cyanobacteria was created by comparing the 16S rRNA gene and cpcBA-IGS sequences to corresponding cyanobacterial sequences available in databases and the sequences obtained in this study for Spirulina laxissima SAG 256.80 and Oscillatoria sancta NIER 10027.
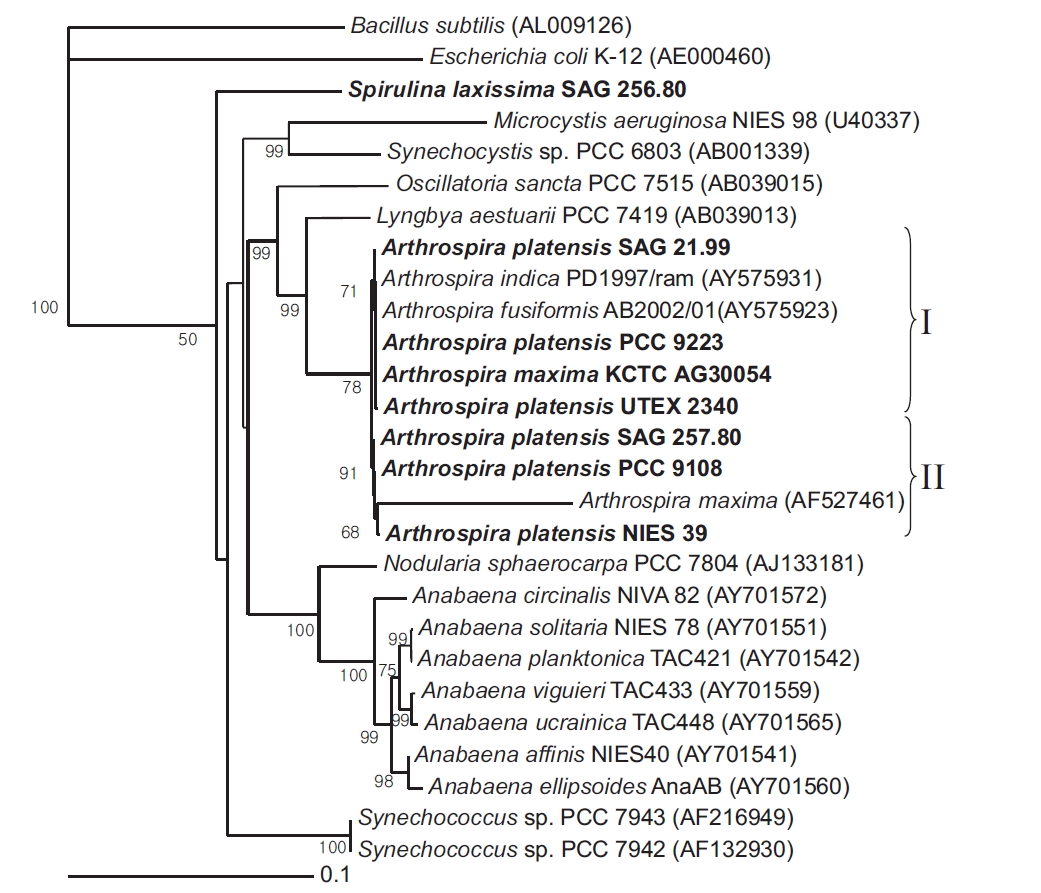
Near-complete 16S rRNA gene sequences were determined for 7 Arthrospira strains and 1 Spirulina strain received from culture collections across the world. A phylogenetic tree was then reconstructed using a NJ analysis based on aligning the 8 sequences with Escherichia coli K-12 and Bacillus subtilis as the outgroup (Fig. 1). The corrected sequence alignment, providing the basis of the phylogenetic analyses, corresponded to positions 8-1512 according to the E. coli numbering system and was 1,383 nucleotides (nt) in length after removing all gaps and ambiguous positions.
The cluster analysis resolved the 7 Arthrospira strains into two main genotypic clusters, designated Group I and II. Group I contained A. platensis SAG 21.99, PCC 9223, UTEX 2340, and A. maxima KCTC AG30054, while Group II contained A. platensis NIES 39, SAG 257.80, and PCC 9108. The bootstrap value between the Group I and Group II clusters was 78.4% in the phylogenetic tree, and the 16S rRNA gene similarity was 99.5%. Thus, the clusters were poorly supported by the bootstrap analysis. However, in the 16S rRNA gene sequences for the Arthrospira strains, the number of different nucleotides was less than 7 out of a total of 1,420 nt. In the geographical analysis, the strains in Group I originated from Chad, Namibia, and Kenya in Africa, while the strains in Group II originated from Chad in Africa and Peru in South America. In a previous report, a complete analysis of the dendrogram structure grouped the strains into two well-separated genotypic groups (Viti et al. 1997). The genotypic diversity of several strains attributed to these two species was also previously investigated on the basis of morphological criteria using a very sensitive total DNA restriction profile analysis (Scheldeman et al. 1999). In this case, the strains were also divided into two well-separated genotypic groups.
[Fig. 1.] Neighbour-joining tree showing relationships among Arthrospira strains and other cyanobacteria, inferred from 16S rRNA gene sequence analyses. Bootstrap values >50% are shown. Bacillus subtilis (AL009126) and Escherichia coli (AE000452) were used as an outgroup to define the root of the tree. Bar, 0.1 substitutions per nucleotide position. SAG, Sammlung von Algenkulturen der Universitat Gottingen, Germany; NIES, National Institute for Environmental Studies Collection, Tsukuba, Ibaraki, Japan; PCC, Pasteur Culture Collection of Cyanobacterial Strains, Paris, France; KCTC, Korean Collection for Type Cultures, Daejeon, Korea; UTEX, Culture Collection of Algae at the University of Texas at Austin, Austin, TX, USA.
The similarity of the 16S rRNA genes between the Arthrospira strains and Lyngbya aestuarii PCC 7419 was about 95%. However, the similarity of the 16S rRNA genes between the Arthrospira strains and S. laxissima SAG 256.80 was about 89%. Thus, as shown by the 16S rRNA gene sequences (Nelissen et al. 1996, Ishida et al. 1997), it would seem that L. aestuarii PCC 7419 is also closely related to Arthrospira and a sister to the clade Planktothrix/Arthrospira.
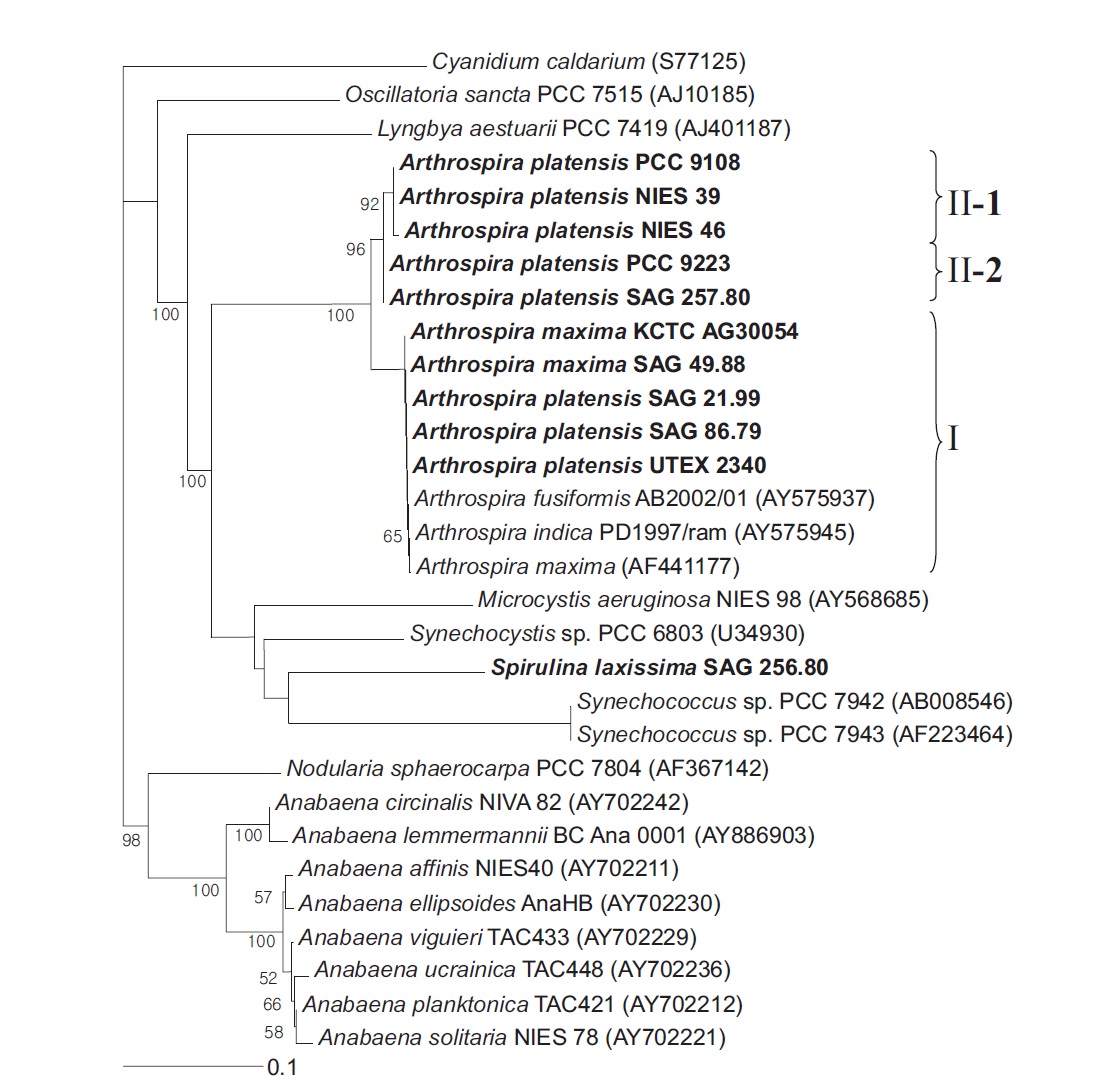
The Arthrospira strain analyses were conducted using both the coding sequences and the spacer. Plus, the outgroup was the cpcBA-IGS from the chloroplast of Cyanidium caldarium. The NJ tree derived from the translated cpcBA-IGS sequences (Fig. 2) clustered the Arthrospira species into two groups, where Group I clustered with the Group II clusters in 100% of the bootstrap trees and the cpcBA-IGS similarity was more than 95.5%. Group I contained A. platensis SAG 86.79, SAG 21.99, UTEX 2340, A. maxima SAG 49.88, and KCTC AG30054, whereas Group II was subdivided into two lineages: one clade containing A. platensis PCC 9108, NIES 39, and NIES 46 supported by 96.6% of the bootstrap replications, and the other clade consisting of A. platensis PCC 9223 and SAG 257.80. The similarity of the strains belonging to Group I was 100%. However, while A. platensis PCC 9223 belonged to Group II-2 based on its cpcBA-IGS sequence, it also belonged to Group I based on its 16S rRNA gene.
With respect to the terminal branching patterns, the results suggested a high degree of congruence between the 16S rRNA genes and the cpcBA-IGS-based phylogenies of the nonmarine members of the picophytoplankton clade, yet with less well delineated subgroups in the more conserved 16S rRNA gene phylogeny (Robertson et al. 2001,
[Fig. 2.] Neighbour-joining tree showing relationships among Arthrospira strains and other cyanobacteria, inferred from cpcBA-intergenic spacer sequence analyses. Bootstrap values >50% are shown. The chloroplast of Cyanidium caldarium was used as an outgroup to define the root of the tree. Bar, 0.1 substitutions per nucleotide position. PCC, Pasteur Culture Collection of Cyanobacterial Strains, Paris, France; NIES, National Institute for Environmental Studies Collection, Tsukuba, Ibaraki, Japan; SAG, Sammlung von Algenkulturen der Universitat Gottingen, Germany; KCTC, Korean Collection for Type Cultures, Daejeon, Korea; UTEX, Culture Collection of Algae at the University of Texas at Austin, Austin, TX, USA.
Crosbie et al. 2003). In a previous report, 16S rRNA gene data and cpcBA-IGS data also indicated that several taxa were genetically heterogeneous and that the taxonomy of cyanobacteria needed to be reconsidered. Manen and Falquet (2002) suggested that 16S rRNA gene data and cpcBA-IGS data were often in agreement. Furthermore, another study investigated the use of the cpcBA-IGS region of the phycocyanin operon, flanked by cpcB and cpcA, as a marker for the subgenus characterization of cyanobacteria (Neilan et al. 1995, Bolch et al. 1996).
The phylogenetic analyses based on the 16S rRNA gene and cpcBA-IGS sequences showed no division between A. platensis and A. maxima, plus the 16S rRNA gene and cpcBA-IGS clusters did not exhibit any well-defined geographical distributions, and instead overlapped in a rather interesting way. Thus, it is now widely accepted that S. laxissima SAG 256.80 is distinct and relatively distant from Arthrospira strains.
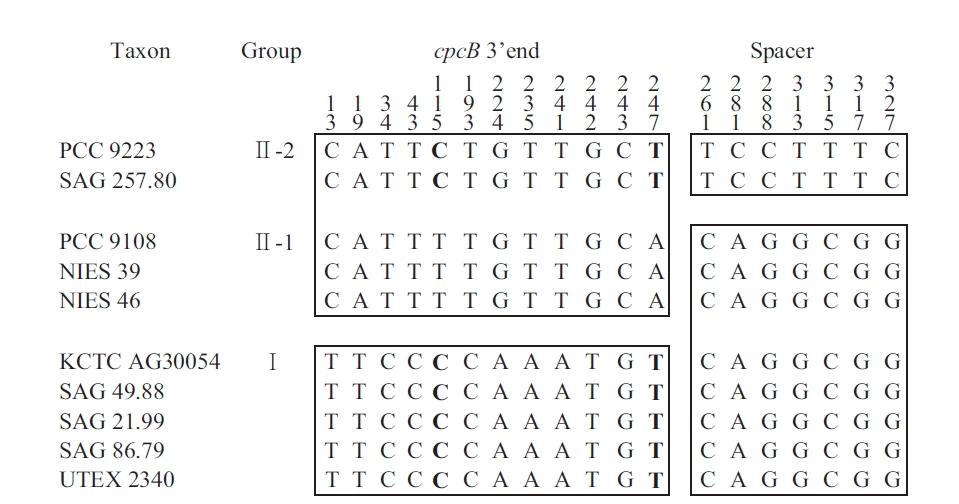
The sequences of the amplified fragments of the genus Arthrospira comprised the last 259 nt of cpcB, a 111 nt spacer, and the first 33 nt of cpcA. These regions were identified based on sequence comparisons using the BLASTN Sequence (http://www.ncbi.nlm.nih.gov/BLAST). For comparison, the Spirulina (Oscillatoriales) sequence comprised 259 nt of cpcB, a 100 nt spacer, and
[Fig. 3.] Mosaic distribution of variable informative sites in cpcBA-intergenic spacer sequence of Arthrospira strains. For easier observation, the strains are classified according to the clusters in Fig. 2. The numbers above the alignment indicate the nucleotide position of the informative sites. Blocks of orthologous sequences are boxed. PCC, Pasteur Culture Collection of Cyanobacterial Strains, Paris, France; SAG, Sammlung von Algenkulturen der Universitat Gottingen, Germany; NIES, National Institute for Environmental Studies Collection, Tsukuba, Ibaraki, Japan; KCTC, Korean Collection for Type Cultures, Daejeon, Korea; UTEX, Culture Collection of Algae at the University of Texas at Austin, Austin, TX, USA.
the first 33 nt of cpcA. For Phormidium autumnale (Oscillatoriales), the cpcB-cpcA locus comprised 224 nt of cpcB, a 89 to 106 nt spacer, and 281 nt of cpcA (Teneva et al., 2005). The sequence analyses of Nodularia isolates (Nostocales) resulted in 283 nt of cpcB, a spacer of 78-79 nt, and 306 nt of cpcA (Barker et al. 1999), whereas for the different Aphanizomenon colonies (Nostocales), this region comprised 227 nt of cpcB, a spacer of 77-103 nt, and 47 nt of cpcA (Barker et al. 2000). For Synechococcus (Chroococcales), the sequence comprised 274 nt of cpcB, a 43 to 60 nt spacer, and 195 nt of cpcA (Robertson et al. 2001). Interestingly, the length of the cpcBA-IGS spacer in Chroococcales was shorter than that in the other cyanobacteria (Nostocales and Oscillatoriales).
The information sites of the cpcB-cpcA DNA matrix for Arthrospira (with the taxa arranged according to the results in Fig. 2) are shown in Fig. 3. The mosaic distribution of the substitutions suggests recombination events at these phycocyanin loci. The first block comprises positions 1-259 (3′ end of cpcB translated sequences), while the second block comprises positions 260-403 (cpcB-cpcA spacer and 5′ end of cpcA translated sequences). Fig. 3 shows the trees obtained using positions 1-259 and 260-403, respectively. While the first block clearly differentiates Groups I and II, as defined in Fig. 3, the substitutions in the second block do not separate Clade 1 and 2, but rather differentiate Group II-2 from all the other strains. In Group II-1 and II-2, the nucleotide substitution was T to C and A to T at 115 and 247, respectively. No amino acid changes were found in any of the groups. However, in Group I and II, the substitutions at 224 and 242 involved a change from Ser to Gly and Cys to Ala, respectively.
The LARD maximum-likelihood method (Holmes et al. 1999) was applied to find the breakpoint in the alignment that gave the highest likelihood under an evolutionary model incorporating recombination. This point was located after position 259, in agreement with Fig. 3. Thus, the Arthrospira tree topology based on the cpcBA-IGS fragment did not reflect the true cladogenesis due to horizontal transfers. Also, there was no correlation between the DNA matrix and the geographical origin. These results were the same as those reported by Manen and Falquet (2002), and also reminiscent of the mosaic distribution of informative sites previously observed in the rbc LX gene of Nostoc (Rudi et al. 1998). In the genus Arthrospira, the phycocyanin locus also indicates intragenic recombination and suggests exchanges of genetic material between strains. Genetic exchanges have also been postulated in Nodularia (Barker et al. 2000) and Phormidium (Teneva et al. 2005).
In summary, the current study supports some previous conclusions based on 16S rRNA gene and cpcBA-IGS sequences that Arthrospira taxa are monophyletic. Thus, the combination of morphological features with molecular markers, such as 16S rRNA gene sequences and cpcBA-IGS sequences, as used in this study, can be useful tools for identification and / or phylogenetic studies at the species level or higher taxonomic ranks in the order Oscillatoriales. When compared with 16S rRNA sequences, cpcBA-IGS sequences may be better suited to resolve close relationships and intraspecies variability. The probe based on cpcBA-IGS that showed a high specificity for cyanobacteria enabled the amplification and sequencing to be simple and fast.