Gelidium Lamour. is composed of approximately 127 described species distributed globally along tropical, subtropical, and artic shorelines (Freshwater and Rueness 1994, Shimada et al. 2000, Millar and Freshwater 2005, Kim et al. 2011, in press, Guiry and Guiry 2012). Members of the genus can be the most abundant organisms within intertidal algal assemblages. Gelidium is economically important as food, and one of the most promising agar sources in rhodophytes. It has recently been used for industrial paper pulp production in Korea (Seo et al. 2010). However, identification of individual Gelidium specimens is notoriously difficult because of the high degree of morphological variation, particularly in the smaller and medium-sized species (Dixon and Irvine 1977a).
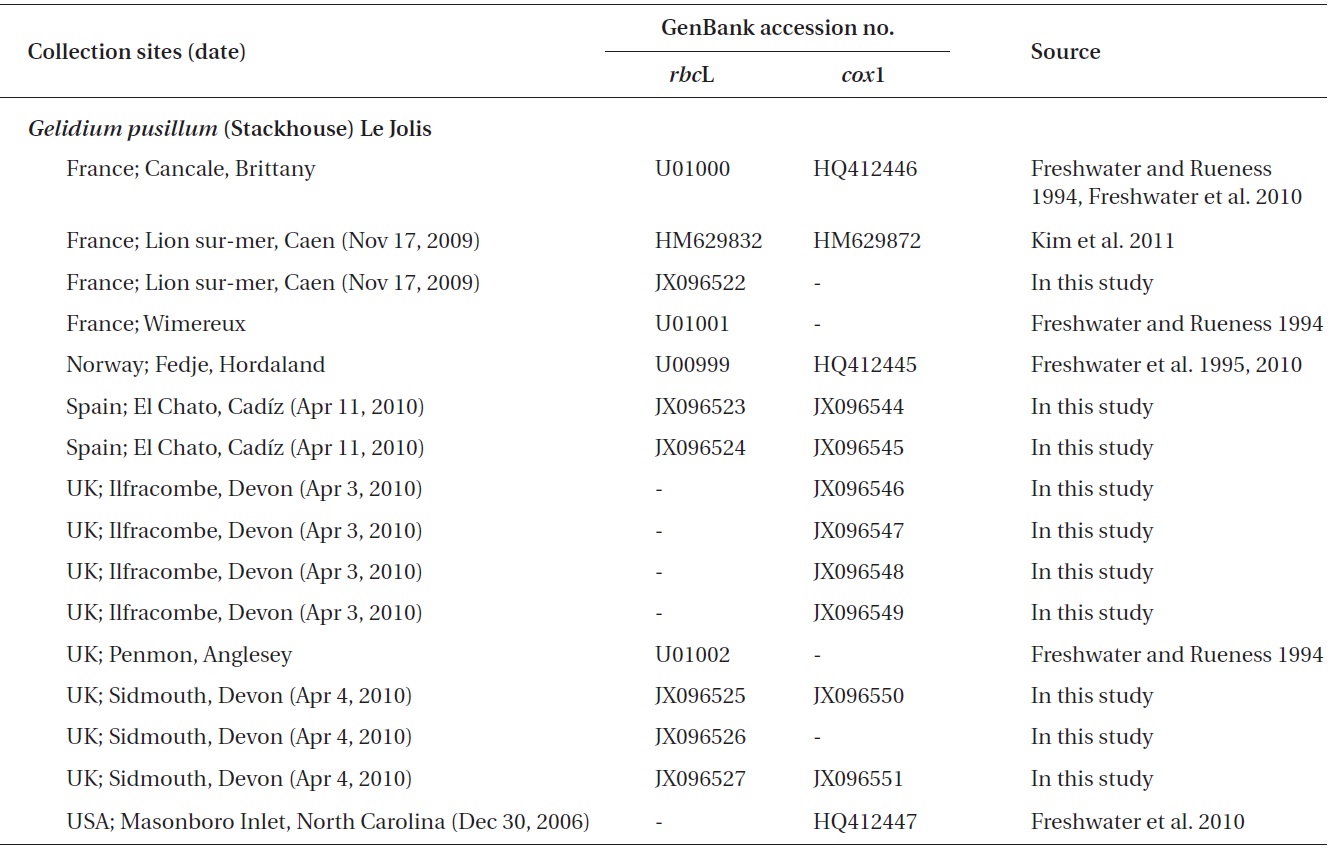
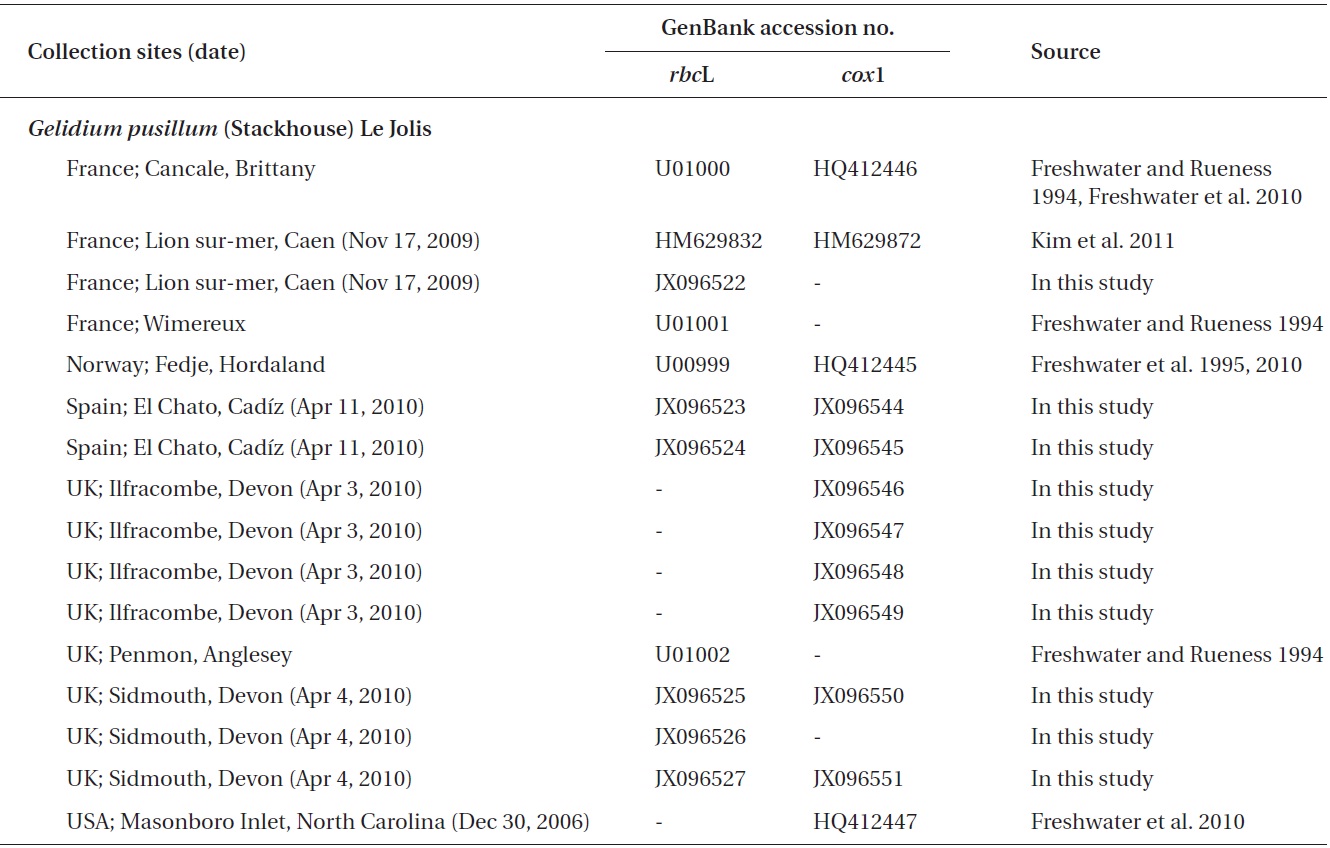
Gelidium crinale (Hare ex Turner) Gallion and G. pusillum (Stackhouse) Le Jolis, which are small and morphologically diverse, are among the most difficult species to identify in red algae, and their distributions are unclear. These species are traditionally recognized as two distinct species (Feldmann and Hamel 1936, Silva et al. 1996), and Womersley and Guiry (1994) found a difference between the types of G. crinale and G. pusillum. On the contrary, G. crinale and G. pusillum were merged by Dixon and Irvine (1977a, 1977b). Seven to nine varieties or formas have been described in each of G. crinale and G. pusillum (Silva et al. 1996, Guiry and Guiry 2012). The name G. pusillum is commonly used for any small tuft-forming Gelidium (Silva et al. 1996). Although G. crinale and G. pusillum are considered as the most widely distributed species in the genus AlgaeBase (Guiry and Guiry 2012), the occurrence of both species in many countries should be reassessed.
In this study we characterized two species, namely G. crinale and G. pusillum, using two molecular markers. To evaluate the relationship and distribution of G. crinale and G. pusillum, we analyzed plastid rbcL, and mitochondrial cox1 including type locality materials. Plastid rbcL is commonly used for Gelidium phylogeny (Freshwater and Rueness 1994, Freshwater et al. 1995, Shimada et al. 2000, Millar and Freshwater 2005, Nelson et al. 2006, Kim et al. 2011). Recent studies have revealed that mitochondrial cox1 is useful for both DNA bar-coding of gelidioid red algae and to examine their distribution patterns (Freshwater et al. 2010, Wiriyadamrikul et al. 2010, Kim et al. 2012). In this study, we included published rbcL sequences analyzed from G. crinale type material (Freshwater et al. 2010) and samples collected in the G. pusillum type locality.
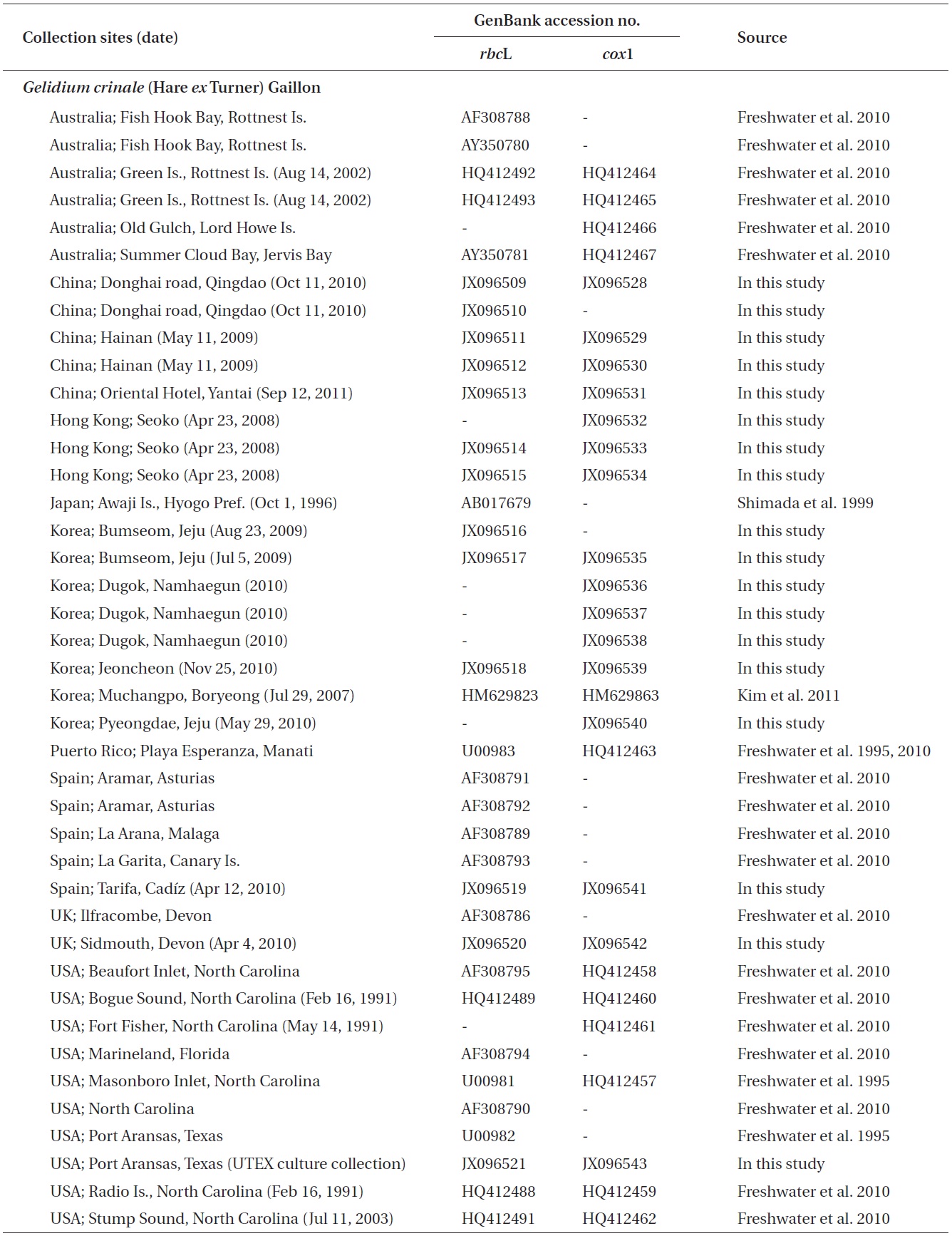
A total of 18 specimens of G. crinale were obtained for this study: 17 collected from 10 locations including China, Hong Kong, Korea, Spain and the UK, and one strain from the culture collection of the University of Texas, UTEX (Appendix A). Ten G. pusillum field collections were made at five locations in France, Spain and the UK. Materials for morphological observations were mounted on herbarium sheets, while clean apical parts of the specimens were desiccated in silica gel for DNA extraction. Tissues were sectioned using a freezing microtome (FX-802A; Coper Electronics Co., Ltd., Kanagawa, Japan), and sectioned preparations were stained with 1% aqueous aniline blue. Photographs were taken with an FX-35DX camera (Nikon, Tokyo, Japan) attached to a Vanox AHBT3 microscope (Olympus, Tokyo, Japan). Voucher specimens were deposited at the herbarium of Chungnam National University (CNUK), Daejeon, Korea.
Twenty-eight specimens were available for DNA extraction (Appendix A). DNA extraction, PCR amplification, and sequencing are described in Geraldino et al. (2010). Primer pairs for amplification and sequencing of each gene were as follows: for rbcL, F7-R753 and F645-RrbcS start (Freshwater and Rueness 1994, Lin et al. 2001, Gavio and Fredericq 2002); and for cox1, cox143F-cox11549R (Geraldino et al. 2006) and C622F-C880R (Yang et al. 2008).
Ninety-three rbcL sequences including 13 new sequences and 65 cox1 sequences including 24 new Gelidium sequences were collated using the Se-Al version 2.0a11 software (Rambaut 1996) and aligned visually. Outgroup taxa used were representatives from Gelidiella Feldmann et G. Hamel, Pterocladia J. Agardh, Pterocladiella Santelices et Hommersand, and Ptilophora (Suhr) Kutzing (Freshwater et al. 1995, Kim et al. 2011).
Maximum likelihood (ML) phylogenetic analysis of rbcL was performed using the GTR + Γ + I model implemented in RAxML software (Stamatakis 2006). We used 200 independent tree inferences with the “number of run” option, with default optimized subtree pruning and regrafting (SPR) rearrangement and 25 distinct rate categories to identify the best tree. Statistical support for each branch was obtained from 1,000 bootstrap replications using the same substitution model and RAxML program settings.
Bayesian analyses (BA) were performed for combined and individual datasets with MrBayes v.3.1.1 (Ronquist and Huelsenbeck 2003) using the Metropolis-coupled Markov chain Monte Carlo (MC3) with the GTR + Γ + I model. For each matrix, one million generations of two independent runs were performed with four chains and sampling trees every 100 generations. The burn-in period was identified graphically by tracking the likelihoods at each generation to determine whether they reached a plateau. The 15,001 trees for rbcL and 22,501 trees for cox1 sampled at the stationary state were used to infer the Bayesian posterior probability.
A statistical parsimony network of cox1 haplotypes was created using TCS version 1.21 software (Clement et al. 2000). Haplotype and nucleotide diversity measurements were performed using DnaSP software (Rozas and Rozas 1999).
A total of 93 sequences from Gelidium and outgroups were aligned using a 1,266-nucleotide (nt) portion of rbcL. Variable sites were found at 492 portions (38.9%), and 393 portions (31%) were parsimoniously informative. All GenBank accessions of G. crinale from nine countries formed a single monophyletic group with maximum
support (Fig. 1). The G. crinale clade consisted of three subgroup; Asian, Australian, and European / American group. Piarwise divergence of G. crinale was up to 2.74%. G. crinale was sister to the clade of G. coulteri Harvey and G. capense (S. G. Gmelin) P. C. Silva (93% for ML and 1.0 for BA). Eleven of G. pusillum sequences from France, Norway, Spain, and UK formed a monophyletic group (100% for ML and 1.0 for BA).
A total of 65 sequences from Gelidium and three outgroups were aligned using a 1,200 nt region of the cox1 gene. Among the 447 (37.3%) variable sites, 400 portions (33.3%) were parsimoniously informative. The topology based on cox1 sequences was congruent with the rbcL phylogeny (Fig. 2). The cox1 ML tree showed that all G. crinale from eight countries were monophyletic (99% for ML and 1.0 for BA). Twelve G. pusillum from France, Norway, Spain, UK, and USA were monophyletic with maximum support.
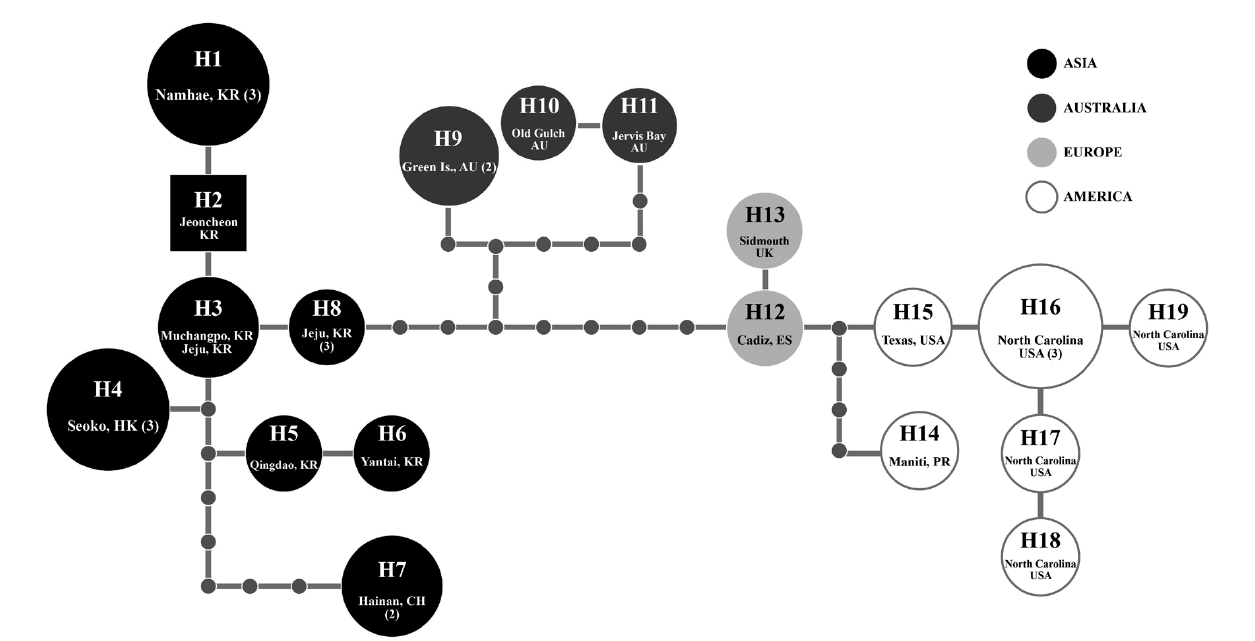
Because of short sequences of cox1 in GenBank, in haplotype analyses, we used 618 nt cox1 fragment of G. crinale from 28 individuals collected in Australia, China, Hong Kong, Korea, Puerto Rico, Spain, UK, and USA. A total of 19 haplotypes among 33 polymorphic sites (5.3%) were found. Haplotype and nucleotide diversities of cox1 within G. crinale were 0.912 ± 0.049 (H) and 0.014 ± 0.005 (π), respectively. The 19 haplotypes were placed in three geographically distinct groups; Asian, Australian, and European / American groups (Fig. 3). All haplotypes in Asia were closely related. However, H7 from Hainan, China was linked to H5 (with six missing haplotypes) and to H3 and H4 (with seven missing haplotypes). Haplotypes H9-H11 were found in Australia, and haplotypes H12-H19 were found in Puerto Rico, Spain, UK, and USA.
Eleven G. pusillum sequences from France, Norway, Spain, and UK formed a monophyletic group with maximum support. Pairwise divergence of G. pusillum was 0.24%. Four haplotypes were found from nine individuals of G. pusillum from France, Spain and UK (data not shown). Four specimens from France and UK shared same haplotype. Three short cox1 sequences (450 nt long) from NCBI (HQ412445-7) from France, Norway and USA were also dentical.
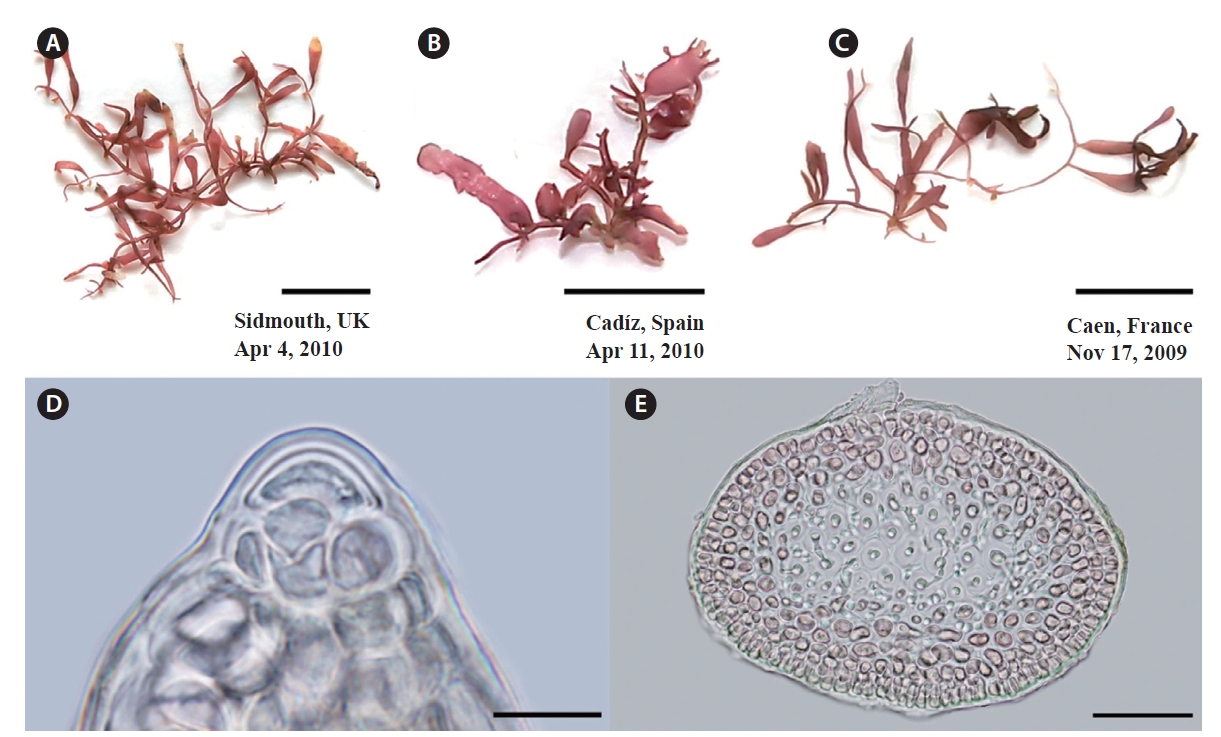
Specimens of G. crinale confirmed by cox1 and rbcL are shown in Fig. 4. Thalli (Fig. 4A & B) from Jeoncheon, Korea and Qingdao, China are purple-red, cartilaginous and composed of terete prostrate axes and erect axes (up to 2 cm high). Specimen from Seoko, Hong Kong (Fig. 4C) are terete to flattened distally (up to 550 μm wide). Spanish specimen appears to have cylindrical branches (Fig. 4D). Large dome-shaped apical cells are evident at the apices and project over the cortical margin (Fig. 4E). Axes and branches consist of cortex and medulla (Fig. 4F). Three to five layered cortex consist of small, pigmented, rectangular surface, and oval inner cortical cells. Medulla is composed of large, colourless medullary cells that were intermixed with internal rhizoidal filaments.
G. pusillum thalli are cartilaginous and prostrate axes are terete, quite regular in diameter, and erect branches (Fig. 5A-C). Erect axes arise from the dorsal sides of indeterminate, prostrate axes, are compressed and up to 1.5 cm in height. Branches develop irregularly branching of up to three orders. Dome-shaped apical cells are present at the apices (Fig. 5D). Axes and branches consist of cortex and medulla (Fig. 5E). The cortex consists of three to four layers of small pigmented cells, whereas the medulla is composed of large colorless cells.
Based on both mitochondrial cox1 and plastid rbcL datasets, Gelidium crinale was distinct from G. pusillum and other congeners of the genus. All G. crinale rbcL sequences from East Asia, Europe, Australia and North America formed a monophyletic clade with a published sequence (AF308786) from the type material (Freshwater et al. 2010). G. crinale specimens from East Asia, Australia and North America corresponded to the description by Feldmann and Hamel (1936), and were identified based on the thalli being caespitose and having cylindrical to compressed prostrate and cylindrical erect axes with filiform branches. G. crinale formed a clade with G. capense from South Africa and G. coulteri from USA. It was difficult to identify a synapomorphic characteristic for these three species.
The intraspecific divergence (0.00-2.74%) of G. crinale from Asia, Australasia, Europe and North America is similar to that (up to 2.65%) of the species in previous studies (Freshwater et al. 2010) and that (0.00-2.45%) of Hypnea flexicaulis Yamaighi et Masuda (Geraldino et al. 2006).
Seven varieties or formas have been described in G. crinale; f. luxurians Collins (type locality, Pacific Beach, San Diego Co. California) (Collins et al. 1906), var. lubricum (Kutzing) Hauck, var. spathulatum (Kutzing) Hauck (Adriatic Sea) (Womersley and Guiry 1994), var.
[Fig. 3.] Nineteen cox1 haplotypes (H1-H19) network of Gelidium crinale from 22 localities in Australia, China, Hong Kong, Korea, Puerto Rico, Spain, UK, and USA. Small grey circles correspond to missing haplotypes and the size of each circle is proportional to the number of individuals analyzed. Numerals in parentheses refer to the number of specimens with identical sequences. AU, Australia; CH, China; ES, Spain; HK, Hong Kong; KR, Korea; PR, Puerto Rico; UK, United Kingdom; USA, United States.
corymbosum (Kutzing) Feldmann et G. Hamel (type locality, Mediterranean Sea, Italy) (Feldmann and Hamel 1936), var. perpusillum Piccone et Grunow (Eritrea, Massawa, Italy) (Piccone 1884), var. platycladum W. R. Taylor (type locality, Port Aransas, Texas, USA) (Taylor 1943) and var. polycladum (Kutzing) Hauck. Of these, var. corymbosum, var. platycladum, and var. perpusillum have been accepted taxonomically in AlgaeBase (Guiry and Guiry 2012).
Our cox1 haplotype network revealed geological structure of the G. crinale populations, but pairwise divergence (up to 2.74% in cox1) is in a range of other species, as mentioned in above. It is therefore difficult to conclude whether our molecular data support infraspecific classification or not. Further sampling is necessary for confirming the infraspecific categories.
G. pusillum specimens from France, Norway, Spain, and USA formed a monophyletic clade with those collected from the type locality (Sidmouth, Devon, England) in both the cox1 and rbcL trees. G. pusillum is characterized by tufted thalli having compressed, oval, or lanceolate branches (Feldmann and Hamel 1936, Silva et al. 1996). All specimens from UK, France and Spain are similar in having compressed thalli and oval or lanceolate branches. The sister relationship of G. pusillum in the genus was not resolved in the cox1 and rbcL trees.
Of nine varieties or formas described within G. pusillum, three are considered as synonyms of the species; var. conchicolum Piccone et Grunow (type locality, Massawa, Eritrea, Ethiopia) (Piccone 1884), f. foliaceum Okamura (Type locality, Shiso-dima, Seto, Kii Province, Japan) (Okamura 1934), and var. minusculum Weber-van Bosse (type locality, Daram Inlet, East coast of Misool Island, Indonesia) (Weber-van Bosse 1921) (see AlgaeBase, Guiry and Guiry 2012). However, six have still been flagged in the AlgaeBase; var. cylindricum W. R. Taylor (type locality, Bahia San Francisco, Ecuador) (Taylor 1945), var. mucronatum P. J. L. Dangeard, var. pacificum W. R. Taylor (Isla Santa Maria, Galapagos Islands, Ecuador) (Taylor 1945), f. pakistancium Afaq-Husain et Shameel (type locality, Gadani, Karachi, Pakistan) (Afaq-Husain and Shameel 1999), var. pulvinatum (C. Agardh) Feldmann (type locality, Cadiz, Spain) (Feldmann and Hamel 1936), and var. simplex P. J. L. Dangeard (type locality, Marocco) (Dangeard 1949).
The monophyly and low genetic variation (up to 0.24%) of G. pusillum reveal that G. pusillum is a species with less genetic diversities and indicate the above six varieties or formas may be synonyms of the species or belong to other species. For example, four varieties of G. pusillum (e.g., var. conchicola, var. cylindricum, var. pacificum, and var. pulvinatum) reported in Korea (Lee 1994, Lee and Kim 1995) have not been found in our recent studies despite many trips in their collection sites (Kim et al. 2011,
in press). Instead, at least one new species in Korea are likely previously misidentified as variant of G. pusillum (Kim et al. in press). Specimen (Fig. 5B) from the type locality of G. pusillum var. pulvinatum, Cadiz, Spain did not reveal variation in molecular data and morphology. Therefore, we suggest that infraspecific classification of G. pusillum may be abandoned.
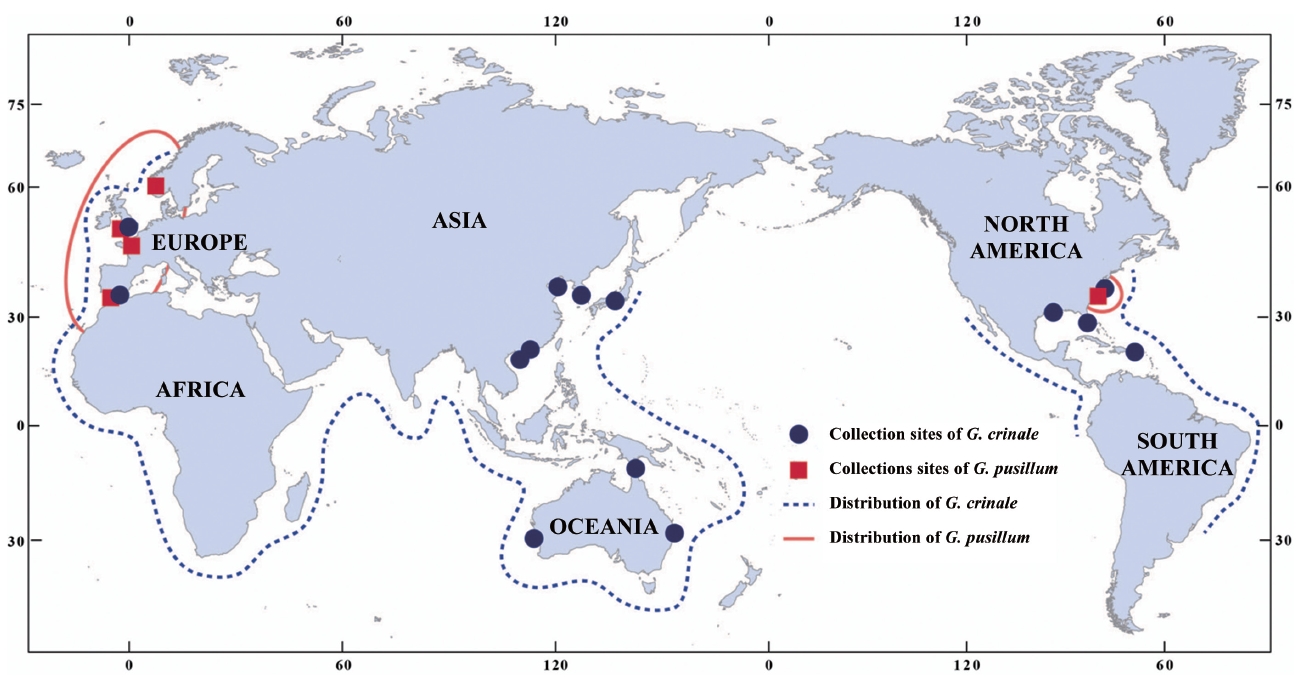
A distribution map of G. crinale and G. pusillum is shown in Fig. 6. Two interesting biogeographic patterns emerge when considering the overall distribution of both species and their distinct lineages revealed by phylogenetic analyses. The parsimony network of cox1 haplotypes revealed a geographic structure (Fig. 3); Asian, Australian, and European / American groups. In the Asian group, seven haplotypes were found and closely related. Australian group is quite distinct from Asian, and European / American groups. Specimens from North Carolina, USA and Europe made one group with a single missing haplotype. The genetic connectivity between Europe and north America may be caused by similar oceanographic conditions. Our results indicate that G. crinale is a cosmopolitan species, although there are high morphological variations and intraspecific divergences. This result is in agreement with previous studies showing that G. crinale is distributed globally (Shimada et al. 1999, Millar and Freshwater 2005, Freshwater et al. 2010)
Contrary to previous studies on its global distribution (e.g., Silva et al. 1996, Guiry and Guiry 2012), our cox1 and rbcL datasets revealed that G. pusillum is likely restricted to Europe and Atlantic North America. Despite basic local alignment search tool (BLAST) search of all sequences registered in GenBank as well as sequences generated in the present study, no accession of G. pusillum from Asia, Australia and Pacific North America matched those from Europe. However, G. pusillum specimens from Spain and France formed a clade with those from Sidmouth, UK. We suggest that records of G. pusillum from Europe and Atlantic North America may be the result of misidentifications, and the specimens filed under G. pusillum should be treated with caution. We agree with the views of Millar and Freshwater (2005) that G. pusillum, reputed to be cosmopolitan (see Silva et al. 1996, Guiry and Guiry 2012), may be limited to areas around Europe and Atlantic North America.
Identifications of G. crinale and C. pusillum based on morphology alone is problematic and usually requires molecular analyses for confirmation. Thus, reports of G. pusillium outside from of Europe and Atlantic North America should be treated with caution, and herbarium specimens identified as G. pusillum in East Asia, Australia and North America should be re-examined. Although G. crinale is common in Europe and other continents, its morphology has been less studied than that of G. pusillum (Rueness and Fredriksen 1990, 1998, Fredriksen et al. 1994). Thus, further investigation of the morphology and phenology of G. crinale is required.




![Maximum likelihood tree of Gelidium using 93 rbcL sequences calculated using the GTR + Γ + I evolution model (-lnL = 10511.506124; substitution rate matrix RAC = 0.951940, RAG = 5.742494, RAT = 1.371355, RCG = 1.258690, RCT = 10.348932, RGT = 1; shape parameter [α] = 1.575834). Maximum likelihood bootstrap values and Bayesian posterior probabilities are shown for each clade. Only bootstrap values ≥50% and ≥0.95 Bayesian posterior probabilities are shown.](http://oak.go.kr/repository/journal/11245/JORHBK_2012_v27n2_83_f001.jpg)
![Maximum likelihood tree of Gelidium using 65 cox1 sequences calculated using the GTR + Γ + I evolution model (-lnL = 9341.027999; substitution rate matrix RAC = 1.153854, RAG = 10.881554, RAT = 1.077417, RCG = 0.000017, RCT = 20.342285, RGT = 1; shape parameter [α] = 1.101521). Maximum likelihood bootstrap values and Bayesian posterior probabilities are shown for each clade. Only bootstrap values ≥50% and ≥0.95 Bayesian posterior probabilities are shown.](http://oak.go.kr/repository/journal/11245/JORHBK_2012_v27n2_83_f002.jpg)