Some echinoids were collected from the coast of Gangwon-do during the period from November 2008 to July 2011 and were identified on the basis of morphological characteristics and molecular analysis of cytochrome oxidase subunit I mitochondrial DNA. Among them, Strongylocentrotus pallidus (Sars, 1871) belonging to the family Strongylocentrotidae of the order Camarodonta is reported for the first time in Korea and is redescribed. The genetic differences ranged from 0.038 to 0.139 between S. pallidus and four other species of genus Strongylocentrotus, but ranged from 0.002 to 0.005 between Korean specimens and GenBank data of S.pallidus. This species is widely distributed in cold sea water along the western part of the North Pacific and the Northwest Atlantic.
The genus
>
Sample collection and identification
The specimens of
>
DNA amplification and sequencing
Genomic DNA was extracted from the gonad tissues of echinoids using by DNeasy blood and tissues kit (Qiagen, Hilden, Germany) and the COI gene was amplified using primers of Knott and Wray (2000): ECO1a (5′-ACCATGC AACTAAGACGATGA-3′) and ECO1b (5′-GGTAGTCTGAGTATCGTCGWG-3′). PCR amplification chemistry con-taining 1.5 μL of genomic DNA, 2.5 μL of 10× PCR buffer (contained MgCl2), 1.0 μL of 2.0 mM dNTPs and each pri-mer, 0.3 μL of nTaq DNA polymerase (Enzynomics, Seoul, Korea) and add up to 25.0 μL with distilled water, and the following conditions: initial denaturation of 2 min at 95℃, 30 cycles of 95℃ 30 sec; 52℃ 1 min; and 72℃ 1 min and a final elongation of 7 min at 72℃. DNA fragments were sequ-enced on an ABI 3730XL sequencer (Applied Biosystems Inc., Forster City, CA, USA) using the ABI Prism Bigdye Terminator v3.1 (Applied Biosystems Inc.).
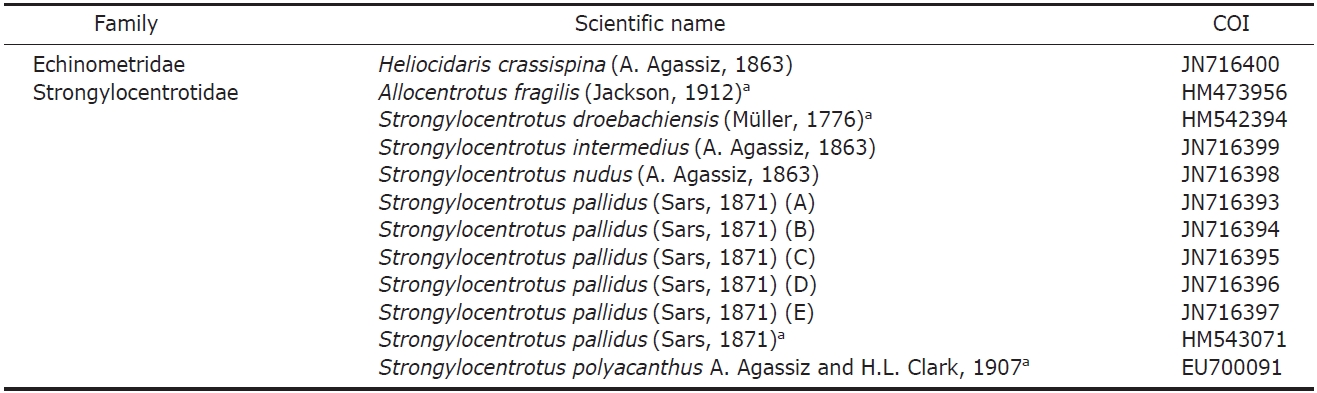
The mitochondrial COI gene was mostly sequenced for this study, but the sequences data of four species which are not distributed in Korea obtained from GenBank (Table 2). COI
[Table 1.] Examined materials of Korean echinoids
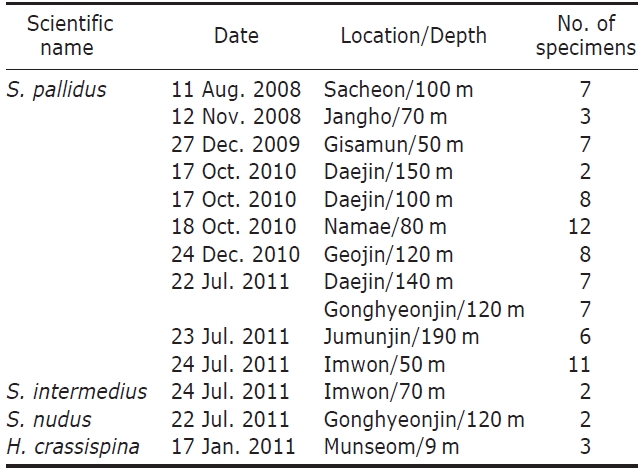
Examined materials of Korean echinoids
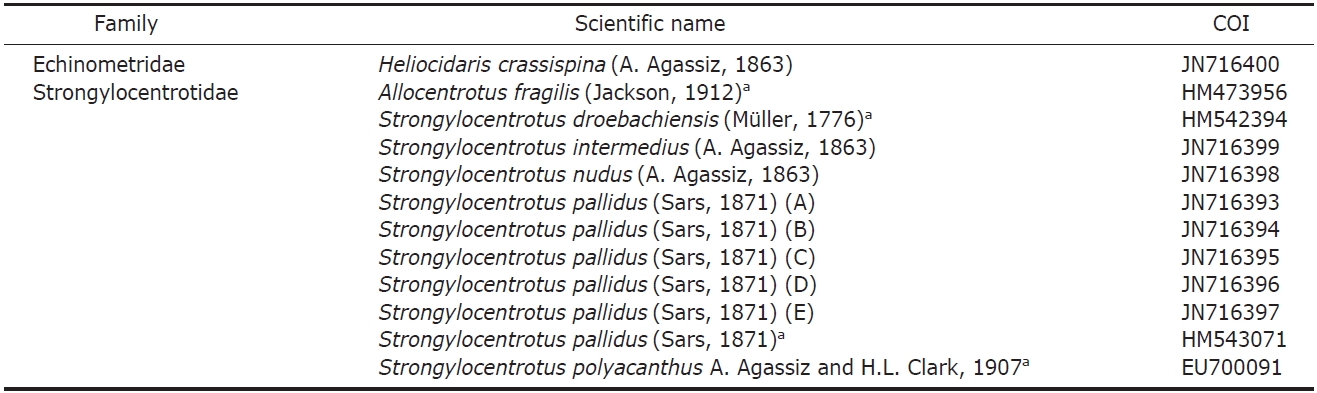
List of taxa included in this study higher taxonomic placement and GenBank accession numbers for their gene sequences
sequences were checked and aligned using by BioEdit v.7.0 (Hall, 1999) and genetic distances were calculated accord-ing to the Kimura 2-parameter model (Kimura, 1980) using by MEGA5 (Tamura et al., 2011). The phylogenetic rela-tionship of the samples was drawn by using the neighbor-joining (NJ), maximum-likelihood (ML) and Bayesian infer-ence (BI). The NJ tree (Saitou and Nei, 1987) was inferred from Kimura 2-parameter genetic distance with bootstrapp-ed 1,000 times using by MEGA5, and the ML analysis with the GTR+G model, determined with jModeltest 0.1.1 (Guin-don and Gascuel, 2003; Posada, 2008), with 1,000 bootstrap replications using by PhyML v3.0 (Guindon and Gascuel, 2003) also BI analysis with same model and analyzed by MrBayes 3.1 with 1×106 generation repeats with the nu-cleotide model 4by4, Nst=6, rates=gamma, Ngammacat=6, and Burnin=2.5×105 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003; Ronquist et al., 2005).
Korean name: 1*연약둥근성게 (신칭)
Class Echinoidea Leske, 1778
Subclass Euechinoidea Bronn, 1860
Order Camarodonta Jackson, 1912
Infraorder Echinidea Kroh and Smith, 2010
Family Strongylocentrotidae Gregory, 1900
Genus Strongylocentrotus Brandt, 1835
1*
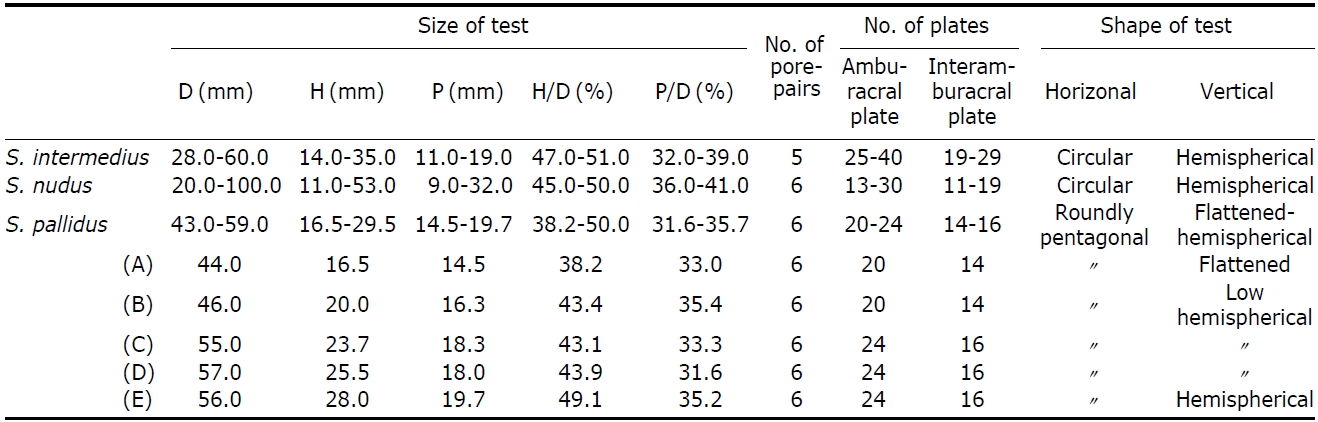
[Table 3.] Molphological characteristics of species of Strongylocentrotus in Korea
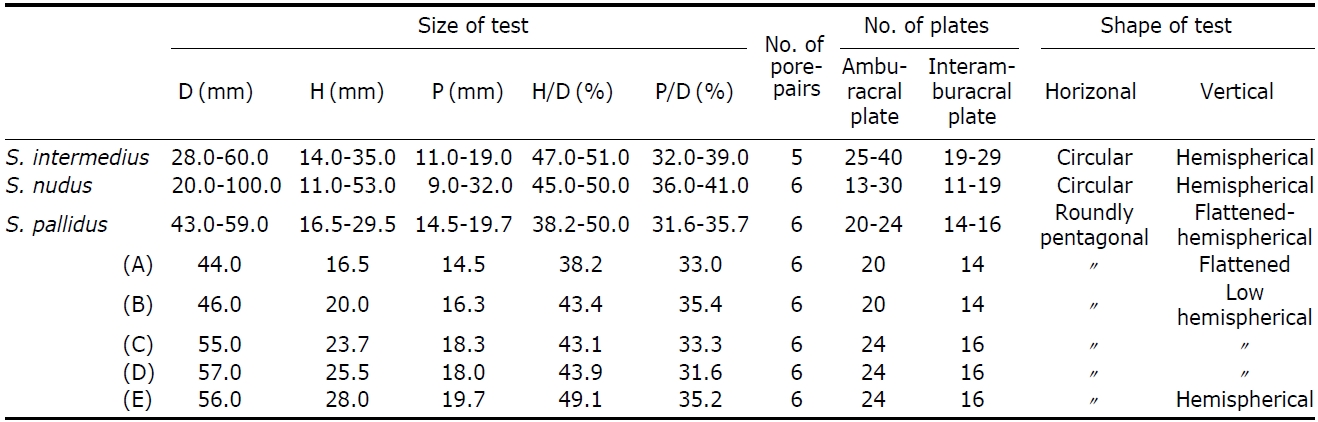
Molphological characteristics of species of Strongylocentrotus in Korea
Key to the species of genus Strongylocentrotus in Korea
1. Ambulacral pore-pairs five in number ?????? S. intermedius Ambulacral pore-pairs six in number ??????????????????????????????????? 2
2. Primary spines long and stout ??????????????????????????????????? S. nudus Primary spines short and not stout ????????????????????? S. pallidus
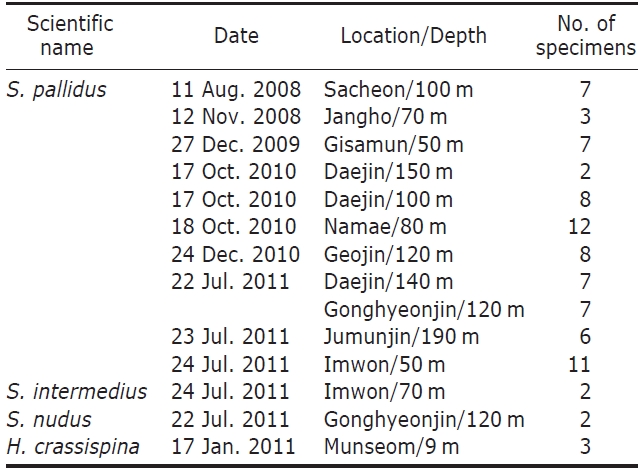
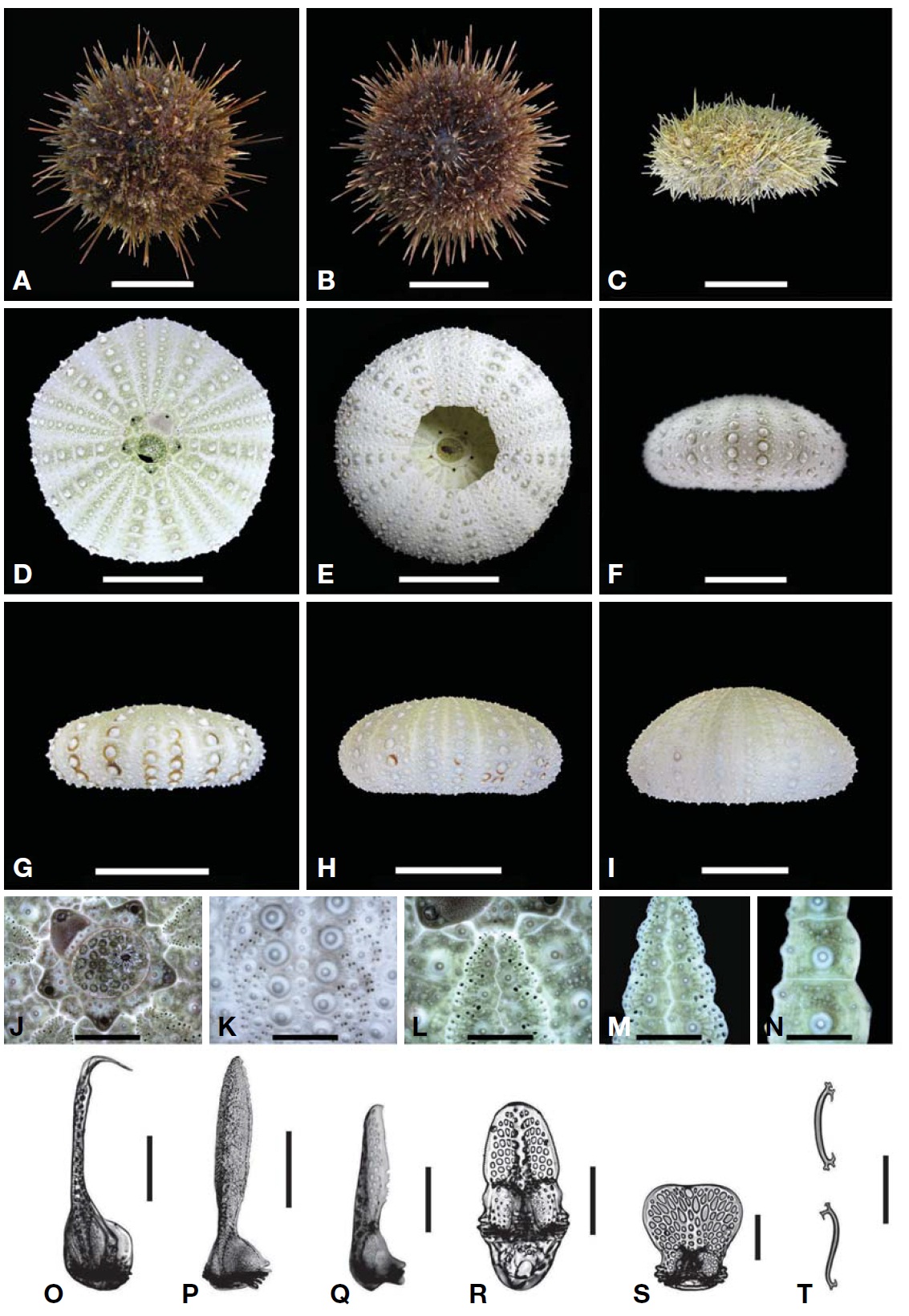
Description.Test form vertically very variable (Table 3); flattened (Fig.1 G), low-hemispherical (Fig.1F, H) and hemi-spherical forms (Fig.1 I). Outline of test roundly pentagonal forms (Fig.1 D). Margin of oral side slightly sunken towards peristome. Test of largest specimen 59 mm in diameter. Six ambulacral pore-pairs presented in an erected arc (Fig.1 K-M). Madreporite has convexed form, larger than genital plates (Fig.1 J). Ocular plates composed five plates, three plates has pentagonal form, stuck between genital plates, like wedges, the other two plates have hexagonal form, situ-ated between genital plates, bordering surnal parts. Surnal parts consist of numerous small plates, with anus situated beside center, surrounded with small blunt spines (Fig.1 J). Globiferous pedicellaria has single slender apical tooth, without lateral tooth (Fig.1 O). Tridentate pedicellaria has two different sizes, large and small form; large ones about five times larger than small one (Fig.1 P, Q). Ophiocephal-ous pedicellaria with fountain pore-pattern valves (Fig.1 R). Triphyllous pedicellaria apple-like shaped, with upraised radial pore-pattern (Fig.1 S). Spicule of tube-foot usually elongated arch form, rarely twisted form, which has trifur-cated end, of which inside node slender, tapered to tip (Fig.1T).
Distribution. Korea (Gangwon-do), Japan (Siaukhu Bay), Bering Sea, Okhotsk Sea, North Pacific (Kuril island, Sakah-lin), Northwest Atlantic (Norway, U.K.).
DNA sequence features. A total of 1,214 base pairs (bp) of the COI mtDNA were obtained from five specimens of
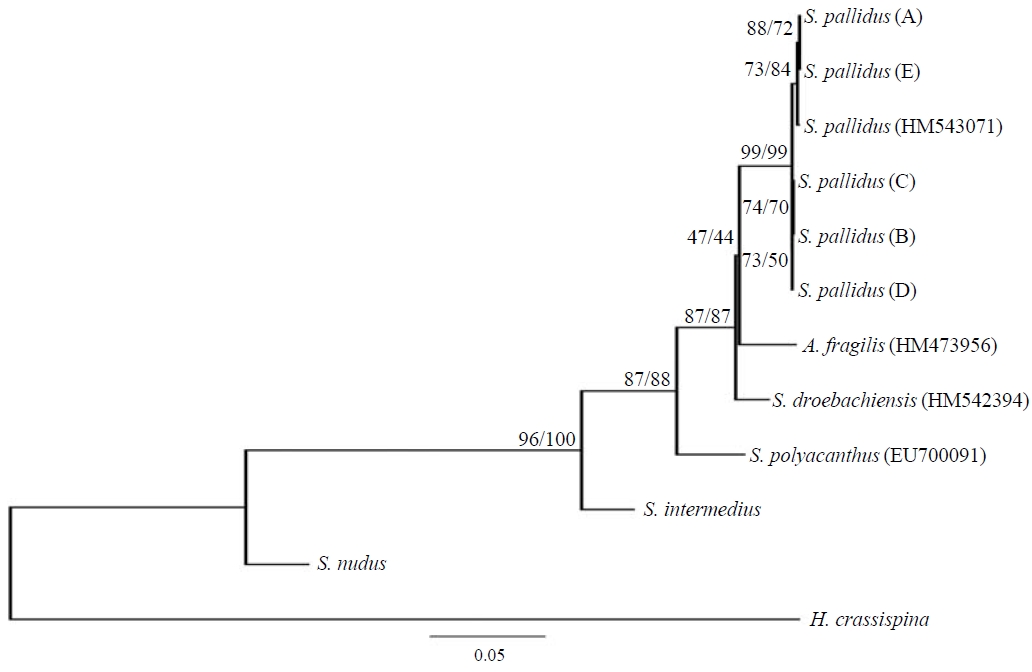
Phylogenetic tree. The phylogenetic relationships of the COI mtDNA from
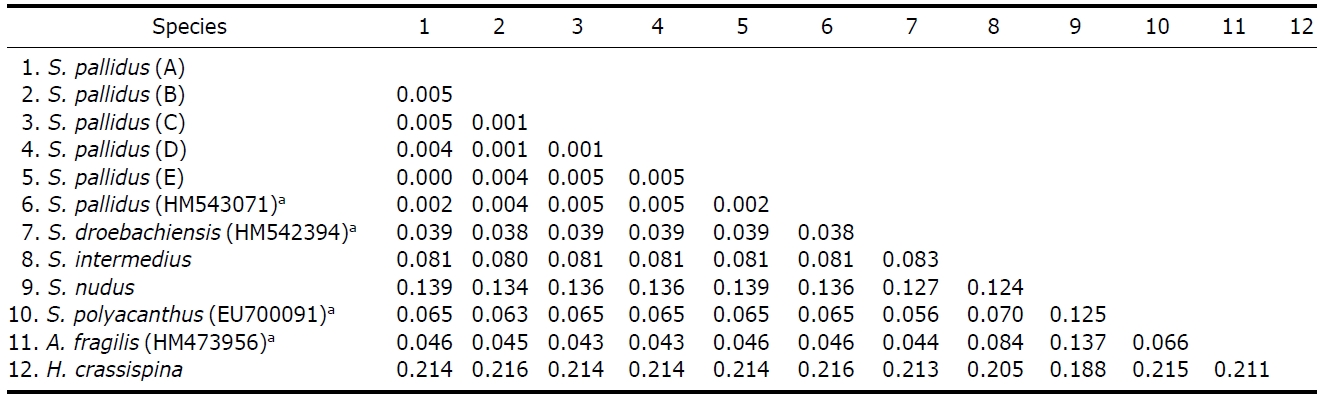
Genetic distances. Kimura 2-parameter genetic distance between
(GenBank) ranged from 0.002 to 0.005 (Table 4). Pairwise(
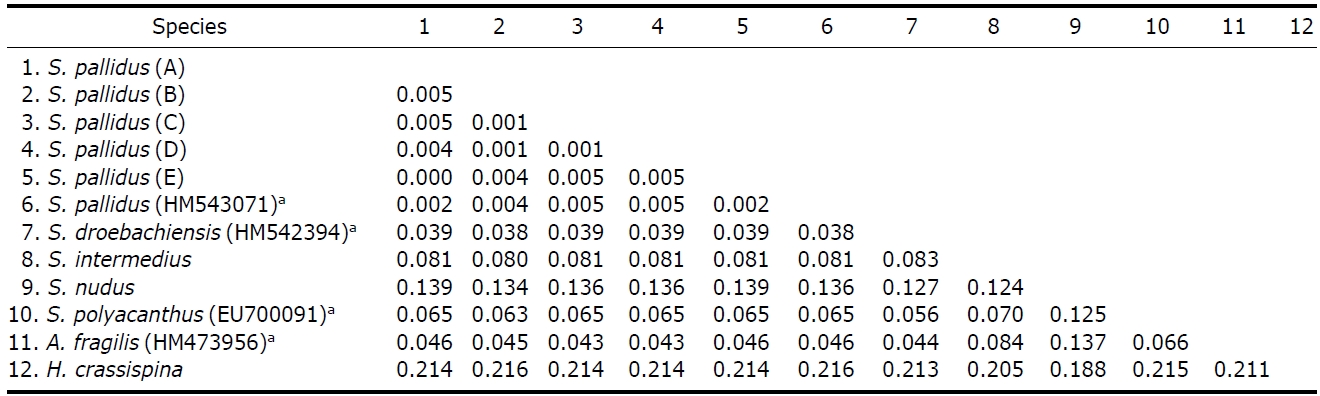
Interspecific pairwise (p) distance values among six species of genus Strongylocentrotus with an outgroup (Heliocidariscrassispina) based on partial sequences of mtDNA COI gene which determined by the Kimura 2-parameter model
tance (=0.039) was lower than the
Two species of
Mortensen (1943) reported that