The tiger population that once inhabited the Korean peninsula was initially considered a unique subspecies (Panthera tigris coreensis), distinct from the Amur tiger of the Russian Far East (P. t. altaica). However, in the following decades, the population of P. t. coreensis was classified as P. t. altaica and hence forth the two populations have been considered the same subspecies. From an ecological point of view, the classification of the Korean tiger population as P. t. altaica is a plausible conclusion. Historically, there were no major disper-sal barriers between the Korean peninsula and the habitat of Amur tigers in Far Eastern Russia and northeastern China that might prevent gene flow, especially for a large carnivore with long-distance dispersal abilities. How-ever, there has yet to be a genetic study to confirm the subspecific status of the Korean tiger. Bone samples from four tigers originally caught in the Korean peninsula were collected from two museums in Japan and the United States. Eight mitochondrial gene fragments were sequenced and compared to previously published tiger subspecies’ mtDNA sequences to assess the phylogenetic relationship of the Korean tiger. Three individuals shared an identical haplotype with the Amur tigers. One specimen grouped with Malayan tigers, perhaps due to misidentification or mislabeling of the sample. Our results support the conclusion that the Korean tiger should be classified as P. t. altaica, which has important implications for the conservation and reintroduction of Korean tigers.
The tiger,
From the Joseon Dynasty to the Japanese colonial period, tigers were seen as a threat to the Korean people and there was a lot of pressure to eliminate tigers from the wild (Endo, 2009; Kim and Lee, 2011). As a consequence, tigers that had once roamed the forests of the Korean peninsula became
[Table 1.] Samples used in this study and haplotypes of the mitochondrial DNA
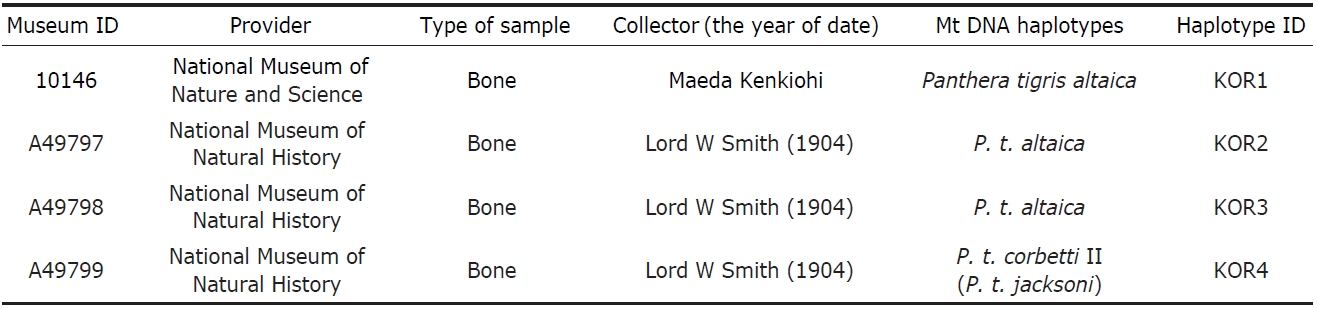
Samples used in this study and haplotypes of the mitochondrial DNA
extinct and the only surviving population in the extreme northeastern Asia is the Amur or Siberian tigers in Russian Far East and northeast China.
The tiger is an animal of great significance in Korea. It is deeply embedded in the Korean history and culture with references found in numerous myths, folk tales, art work, and old sayings. Although extinct in the wild, the tiger lives on in the minds of the Korean people through the use of tiger symbols in logos and signs that represent Korea. Currently, the lack of tigers as a top predator has resulted in an uncon-trolled population growth of large ungulates such as deer and wild boar causing both property damage and injuries around the country. Despite the continued attention to the tiger in Korea, there is a lack of scientific data to understand the evolutionary history of the species in this region.
There has been significant debate as to whether the tigers that once inhabited the Korean peninsula are indeed the same subspecies as the tigers that survives in the Russian Far East. In the early 20th century, Brass (1904) classified the Korean tiger as
Korean tigers were extirpated from the wild and some of the remains such as bones and skins were scattered around the world. To acquire specimens with reliable records on cap-ture locality, collector, and year of capture, an intensive sur-vey of known collections was conducted. Four specimens were found from two museums; one in the National Science Museum, Tokyo (NSMT), Japan and three in the National Museum of Natural History (NMNH) in Washington, DC, USA (Table 1). The information regarding their origin was crosschecked using several independent references. Chung(1978) reported that an American doctor, William Lord Smith, while visiting Korea caught wild boar, roe deer, and three tigers near Mokpo in 1902. In addition, there is a record that Dr. Smith hunted three tigers from the southwestern peninsula of Korea; two tigers by himself and one by his hun-ter (Hollister, 1912). According to the records of the NMNH, Smith collected samples from Korea in 1904 then donated the specimens to the NMNH in 1906. Even though there is a conflict between the times of specimen collection, all records indicate that the three tigers in the NMNH were captured by Smith in Korea. The single specimen in the NMNS was taken over from Tokyo museum in 1989, which was initially col-lected by Maeda Kenkichi who lived in Korea from 1878 to 1887 (Dajokan, 1880).
DNA extraction was carried out in a dedicated ancient DNA laboratory in the Laboratory of Genomic Diversity (LGD), Frederick, USA and replicated at the Conservation Genome Resource Bank for Korean Wildlife (CGRB), Seoul, South Korea. Whole genomic DNA was isolated in multiple extracts using QIAamp DNA Micro Kit (Cat. No. 56304; Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. Multiple extracts from each individual were used to confirm the consistency of ancient DNA from contamina-tion and to facilitate PCR of ancient material. In accordance with the procedure of Driscoll et al. (2009), five mitochon-drial genes (
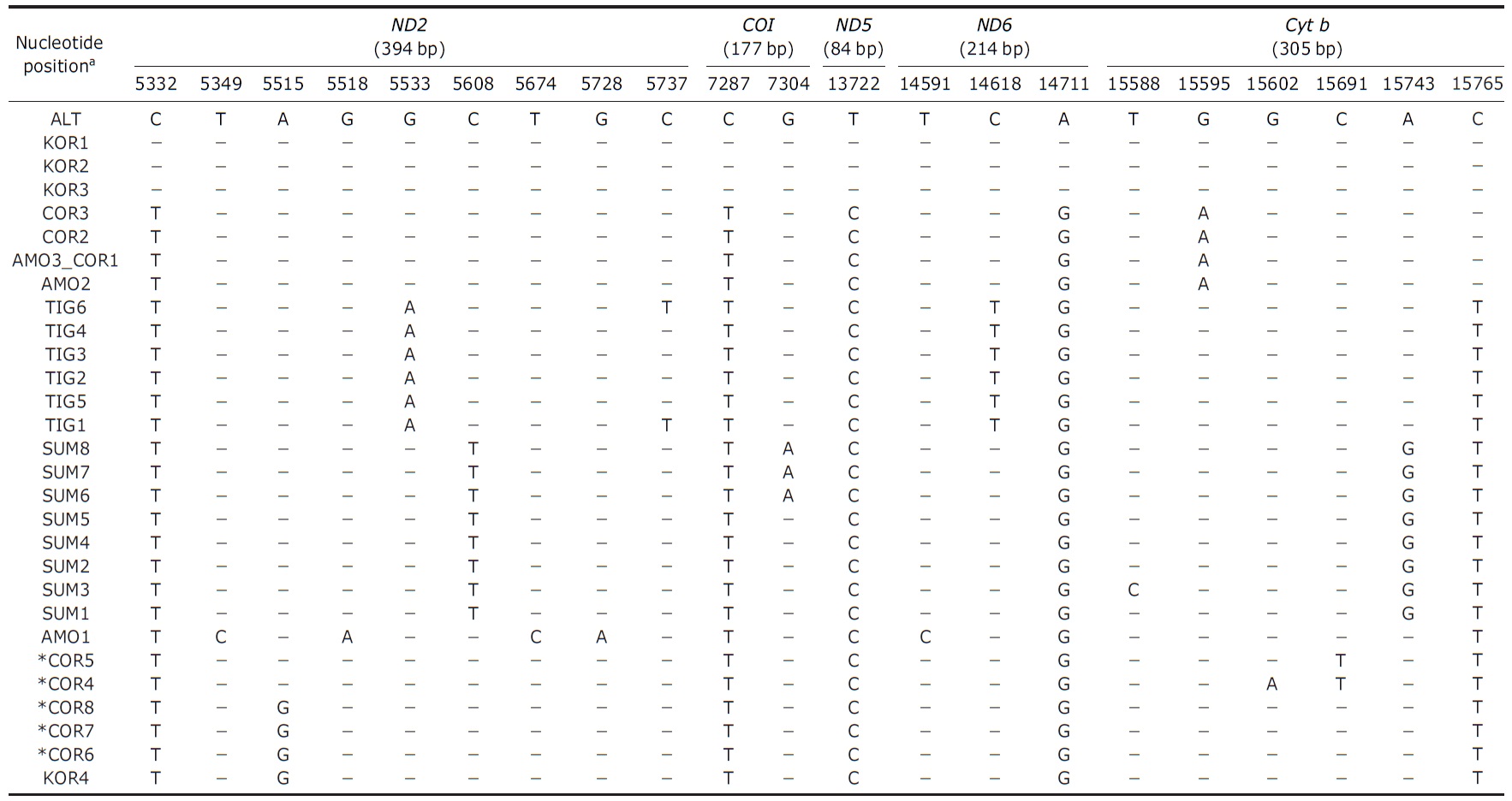
[Table 2.] Polymorphic sites of tigers and mitochondrial gene information used in this study
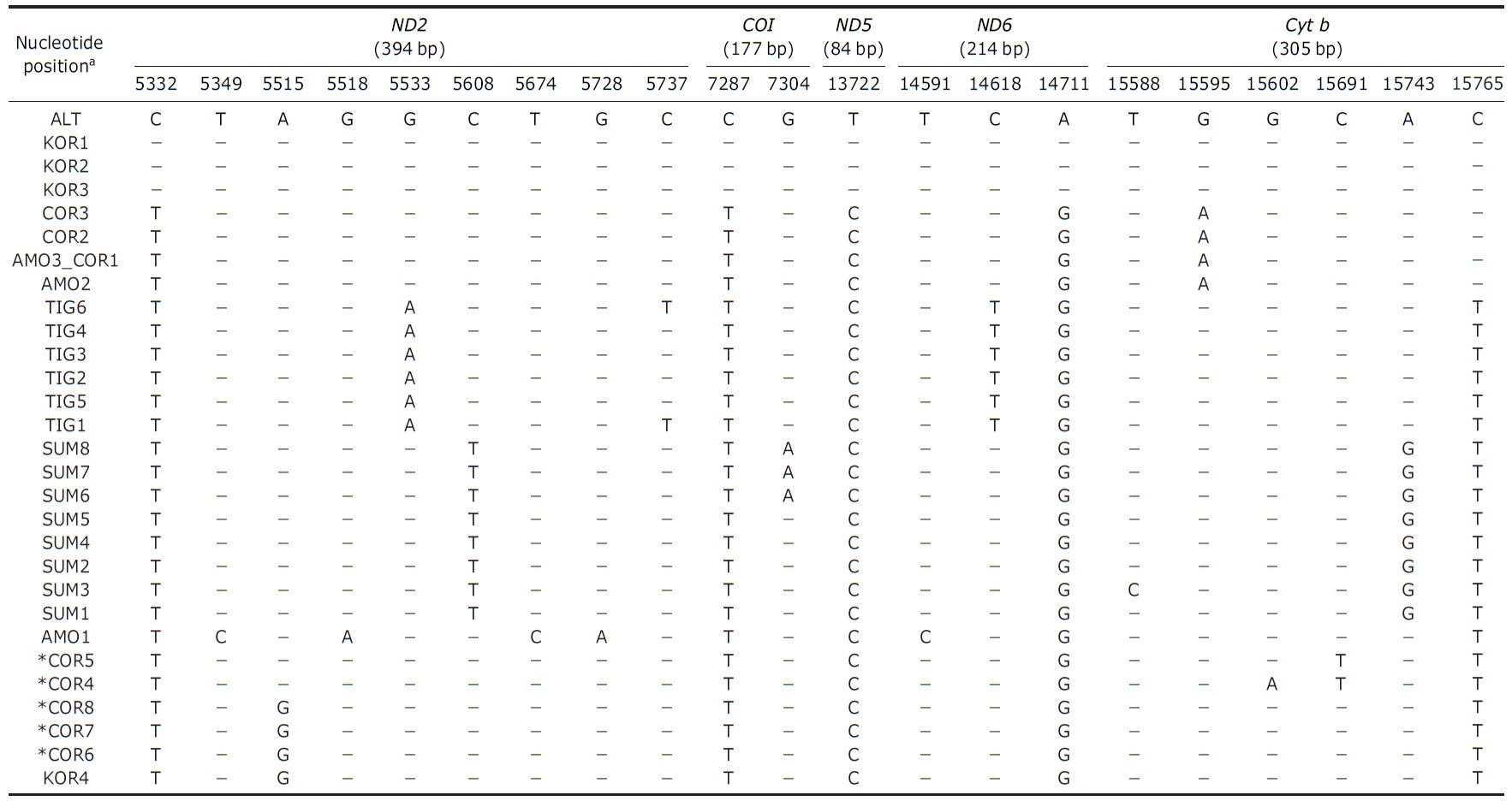
Polymorphic sites of tigers and mitochondrial gene information used in this study
Mitochondrial gene sequences of tiger subspecies from a previous study were retrieved from GenBank for compara-tive phylogenetic analysis (Luo et al., 2004). Sequences were aligned using CLUSTAL_X (Thompson et al., 1997), and further modifications were made by eye using Geneious 5.3 (Drummond et al., 2011). Polymorphic sites were extracted using DNAsp v4.10 (Rozas et al., 2003). The most likely evo-lutionary model parameters were identified using the Akaike Information Criterion (AIC) in Modeltest v3.06 (Posada, 2008). The most suitable model of DNA substitution was HKY+G. An optimal phylogenetic tree was constructed using distance neighbor-joining method in PAUP version 4.0b10 (Swofford, 2002). Eight fragments were combined for a phylogenetic analysis. The robustness of the tree was evaluated by bootstrap resampling (1,000 random replica-tions) (Felsenstein, 1985).
DNA was successfully recovered from bone powders of tigers that were collected in Korea around 100 year ago. Five seg-ments (
DNA were amplified and sequenced from all samples using eight primer sets to yield a total of 1,174 base pairs includ-ing 22 subspecies diagnostic variable sites (Table 2) (Luo et al., 2004).
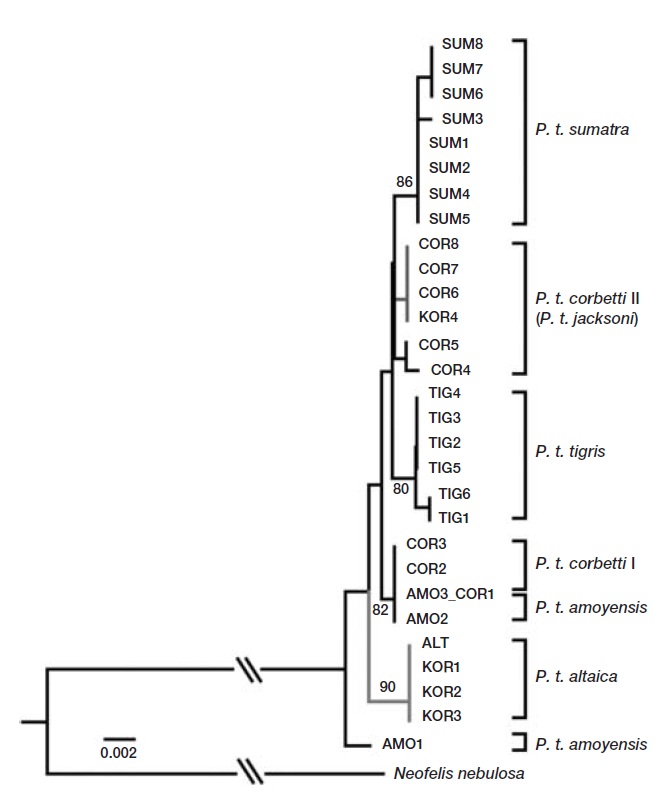
Analysis of the sequences revealed two haplotypes from the four individuals in this study. When compared to previ-ously published sequences, one haplotype shared by three individuals (KOR1 from NMNS; KOR2 and KOR3 from NMNH) was identical to the Amur subspecies,
Tigers are currently listed as an endangered species class I by the Ministry of Environment in South Korea. Although many people will not be convinced that tigers still survive on the peninsula without any scientific evidence, it is broadly believed by the public that they are not extinct. As the Korean tiger disappeared before the genetic techniques were widely employed, it was an unresolved question as to whether to classify the Korean tiger as an independent subspecies,
We identified two mtDNA haplotypes from four 100-year-old specimens of Korean tigers. One haplotype was identical to that of the Amur tiger (
was found in this study because it is highly unlikely that the Malayan tiger inhabited the Korean peninsula a century ago.
The surprising result can be explained by either 1) a mis-handling of the specimens between 1902 or early 1903 when Dr. Smith and his hunter collected the samples and 1906 when they were donated by C. Hart Merriam to the NMNH on behalf of Dr. Smith (database of the NMNH) (Hollister,1912; Chung, 1978), 2) mislabeling during 100 years of stor-age at the museum, or 3) cross-contamination during the expe-riment due to the nature of ancient DNA. The last source has the lowest possibility since all lab work was carried out in a dedicated laboratory for ancient DNA where the DNA of the Malayan tiger was not handled. However, there are reliable records showing that Dr. Smith, the collector of Korean tigers,caught the three tigers in Mokpo, Korea in 1902 or early 1903 and that these tigers were deposited at the National Museum of National History, USA (Hollister, 1912; Chung, 1978). Without further information on the specimens and their sto-rage, it is not possible to verify the precise basis of the ques-tionable results.
In conclusion, except for the individual identified as Mala-yan tigers, the results presented here suggest that the Korean tiger is not an independent subspecies,
The current population size of Amur tigers (