The geographic distribution of the Siberian weasel (Mustela sibirica Pallas, 1773) extends from Siberia through Japan, Korea, and China to northern Thailand (Corbet, 1978), but the subspecies classification of M. sibirica is still unclear; Corbet (1978) reclassified 26 nominal subspecies of M. sibi-rica into seven subspecies (sibirica, manchurica, subhema-chalana, itatsi, sho, quelpartis, and lutreolina). However, Wilson and Reeder (2005) treated the two subspecies (itatsi and lutrolina) as full species of M. itatsi and M. lutreolina, and recognized 12 subspecies, including M. sibirica (canigula, charbinensis, coreanus, davidiana, fontanieri, hodgsoni, manchurica, moupinensis, sibirica, quelpartis, subhemach-alana, and taivana). Three subspecies (M. s. coreanus Doma-niewski from the Korean Peninsula, M. s. quelpartis Thomas from Jeju Island, and M. s. manchurica Brass from northeast-ern China) inhabit Korea and neighboring northeastern China.
In the genus Mustela, molecular phylogenies have been inferred from nuclear DNA interphotoreceptor retinoid bind-ing protein (IRBP) and mitochondrial DNA (mtDNA) cyto-chrome b sequences (Sato et al., 2003), mtDNA cytochrome b and 12S rRNA sequences (Kurose et al., 2008), and three mtDNA loci (Hosoda et al., 2011). The infraspecific diversity of M. sibirica has been examined with 15 cytochrome b sequ-ences from Taiwan, far-eastern Russia, Japan, and Korea, but three haplotypes with partial sequences (402 bp) from three specimens of M. s. coreanus on the Korean Peninsula were used for the analysis (Hosoda et al., 2000).
In this study, we obtained complete mtDNA cytochrome
b sequences (1,140 bp) from 25 specimens of two M. sibirica subspecies in Korea (M. s. coreanus on the Korean Peninsula and M. s. quelpartis on Jeju Island), and these sequences were compared to one corresponding M. s. coreanus haplotype, available from GenBank, to determine the degree of genetic divergence between these two subspecies and to examine the taxonomic status of M. s. quelpartis.
We collected 25 specimens of two subspecies from eight locations in Korea (16 specimens of M. s. coreanus from six locations across the Korean Peninsula and nine specimens of M. s. quelpartis from two locations on Jeju Island) for the cytochrome b sequence analysis, as given in Table 1. Small pieces of muscle were collected and preserved in a deep freezer.
Total cellular DNA was extracted using a genomic DNA extraction kit (Intron Co., Seoul, Korea). The cytochrome b gene was PCR-amplified using the L14724 and H15149 pri-mers, designed by Irwin et al. (1991). The PCR thermal cycle employed was as follows: 94℃ for 5 min; 94℃ for 1 min, 56℃ for 1 min, 72℃ for 1 min (28 cycles); and 72℃ for 5 min. The amplified products were purified using a DNA Prep-Mate kit with a silica-based matrix (Intron Co.) to remove primers and unincorporated nucleotides. The purified PCR products were analyzed with an automated DNA Sequencer (Perkin Elmer, Norwalk, CT, USA) at Bioneer Co. (Seoul, Korea).
We obtained 25 complete cytochrome b sequences (1,140 bp) from two subspecies of M. sibirica in Korea (Table 1), and these sequences were compared to one corresponding sequence of M. s. coreanus from Mt. Jiri (accession no: AB 564135), available from GenBank.
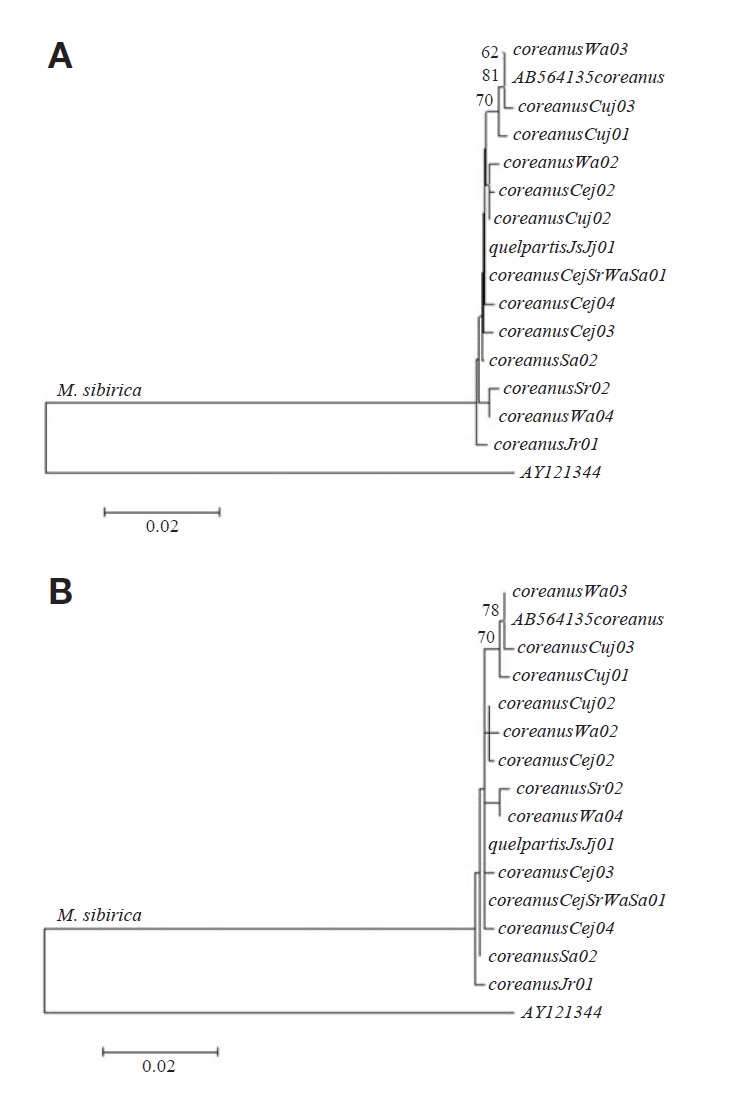
Sequence alignment, detection of parsimonious informative sites, model selection, calculation of nucleotide distances, and tree constructions with 1,000 bootstrapped replications were conducted using MEGA5 (Tamura et al., 2011). The Jukes-Cantor model, showing the lowest Bayesian Information Cri-terion scores, was selected, and neighbor-joining and maxi-mum-likelihood trees were constructed. Martes americana (accession no: AY121344) was used as out group.
Fourteen haplotypes were identified from 25 cytochrome b sequences (1,140 bp) of two Mustela sibirica subspecies in Korea (Table 1). One haplotype (JsJj01) was revealed from nine specimens of M. s. quelpartis at two locations on Jeju Island (four specimens at Seoguipo and five specimens at Jeju), and 13 haplotypes were identified from 16 specimens of M. s. coreanus at six locations on the Korean Peninsula (haplotype Jr01 from Mt. Jiri; CejSrWaSa01 from four loca-tions of Cheongju, Mt. Songri, Mt. Weolak, and Mt. Seolak; Cej02-Cej04 from Cheongju; Sr02 from Mt. Songri; Cuj01-Cuj03 from Chungju; Wa02-Wa04 from Mt. Weolak; and Sa02 from Mt. Seolak). Fourteen cytochrome b haplotypes of two M. sibirica subspecies are deposited in GenBank under accession nos. JQ739185-JQ739198.
Within 15 cytochrome b haplotypes (14 haplotypes from this study and one haplotype from GenBank), 20 sites (1.75%) were variable, and nine sites (0.79%) were parsimonious in-formative. The average Jukes-Cantor nucleotide distance among the 14 haplotypes of M. s. coreanus was 0.46% (range, 0.00 to 0.79%), and, among them, one haplotype (Wa03) from Mt. Weolak was identical to another haplotype (GenBank accession no: AB564135) from Mt. Jiri.
Additionally, one haplotype (JsJj01) of M. s. quelpartis from nine specimens at two locations on Jeju Island was iden-tical to one haplotype (CejSrWaSr01) of M. s. coreanus, from four specimens at four locations on the Korean Peninsula, and the average distance between M. s. quelpartis and M. s.coreanus was 0.26% (range, 0.00 to 0.53%). Neighbor-join-ing and maximum-parsimony trees with 15 haplotypes from two subspecies of M. sibirica are shown in Fig. 1. All 15 haplotypes formed a single clade, indicating that the subspec-ies quelpartis is not genetically distinct from the subspecies coreanus.
Bradley and Baker (2001) noted that a genetic distance of < 2% based on the cytochrome b gene was typical of popula-tion and infraspecific variation. In this study, M. s. quelpartis from Jeju Island was monogenic, and the average distance among 14 haplotypes of M. s. coreanus was 0.46% (range, 0.00 to 0.79%), indicating that genetic divergence within each of the two M. sibirica subspecies, based on the cytoch-rome b gene, was low (< 1%). Further studies with additional specimens are needed to confirm this conclusion.
Johnson et al. (2000) noted that island populations should diverge over time (genetically and morphologically) from respective mainland species populations. In this study of cyto-chrome b sequences (see Fig. 1) two subspecies of M. sibirica formed a single clade and haplotype JsJj01 from nine M. s.quelpartis at two locations on Jeju Island was identical to haplotype CejSrWaSr01 from four M. s. coreanus at four locations across the Korean Peninsula. Average nucleotide distance between the two subspecies was 0.26% (range, 0.00 to 0.53%), indicating that M. s. quelpartis from Jeju Island was not genetically divergent from M. s. coreanus on the Korean Peninsula.
In addition, Lomolino et al. (2010) noted that isolated ani-mals and plants dispersed across formerly submerged land bridges, during the last glacial maxima. Thus, we considered that the lack of divergence between the two M. sibirica subspecies (see Fig. 1) may have resulted from the free disper-sal of the Siberian weasels (M. sibirica) from the Korean Peninsula to Jeju Island through land bridges formed during the last glacial maxima.
However, M. s. quelpartis from Jeju Island is distinct in pelage color from M. s. coreanus on the Korean Peninsula (Thomas, 1906). We found that the results from this cytoch-rome b sequencing study (see Fig. 1) do not support the cur-rent M. sibirica subspecies classification.
A subspecies is an aggregate of phenetically similar popu-lations of a species differing taxonomically from other pop-ulations of that species (Mayr and Ashlock, 1991), and it was advocated that a classification should be the product of all available characters distributed as widely and evenly as pos-sible over the organisms studied (Huelsenbeck et al., 1996). Thus, we conclude that M. s. quelpartis from Jeju Island is a subspecies with only morphological differences.
The shortcomings of using mtDNA sequences for the systematics of hares (genus Lepus) were pointed out, and it has been recommended to include nuclear gene pool evidence
(Slimen et al., 2008). Morphometric and genetic analyses with nuclear and other mtDNA markers from the two M. sib-irica subspecies have not been performed yet, and we pro-pose further systematic analyses with morphometric charac-ters and other DNA markers to confirm the taxonomic status of M. s. quelpartis.