DNA barcoding of Bryozoa or “moss animals” has hardly advanced and lacks reference sequences for correct species identification. To date only a small number of cytochrome c oxidase subunit I (COI) sequences from 82 bryozoan species have been deposited in the National Center for Biotechnology Information (NCBI)GenBank and Barcode of Life Data Systems (BOLD). We here report COI data from 53 individual samples of 29 bryozoan species collected from Korean seawater. To our knowledge this is the single largest gathering of COI barcode data of bryozoans to date. The average genetic divergence was estimated as 23.3% among species of the same genus, 25% among genera of the same family, and 1.7% at intraspecific level with a few rare exceptions having a large difference, indicating a possibility of presence of cryptic species. Our data show that COI is a very appropriate marker for species identification of bryozoans, but does not provide enough phylogenetic information at higher taxonomic ranks. Greater effort involving larger taxon sampling for the barcode analyses is needed for bryozoan taxonomy.
Bryozoans or “moss animals” are distributed around the world and presently more than 5,000 species are known to exist.They are fouling animals, which are easily introduced into alien regions via ships and/or ballast waters (Seo, 2005). The attachment of these animals to either naturally or artificially submerged surfaces, such as ship hulls, enables their frequent relocation. Although bryozoans are taxonomically and ecologically important marine invertebrates, DNA barcoding of them are not yet progressed. Studies based on the standard maker of DNA barcoding of most animal species, mitochondrial cytochrome c oxidase subunit I (COI) sequences, are very rare in bryozoans and confined to a few species with,for instance, concerns of phylogeography (Gomez et al., 2007)and invasion (Mackie et al., 2006).
Approximately, 130 species are known to be distributed in Korea; however, morphological identification of these taxa is difficult due to the lack of distinguishing characteristics,particularly among closely related species. For this reason, we expect that there should be a number of specific synonymy.DNA barcoding, therefore, should contribute significantly to the proper identification of bryozoan species. To date DNA barcodes of only 82 bryozoan species have been deposited in the National Center for Biotechnology Information(NCBI) and the database of Barcode of Life Data Systems(BOLD). Thus, there is an urgent need for augmenting the worldwide database of COI barcodes.
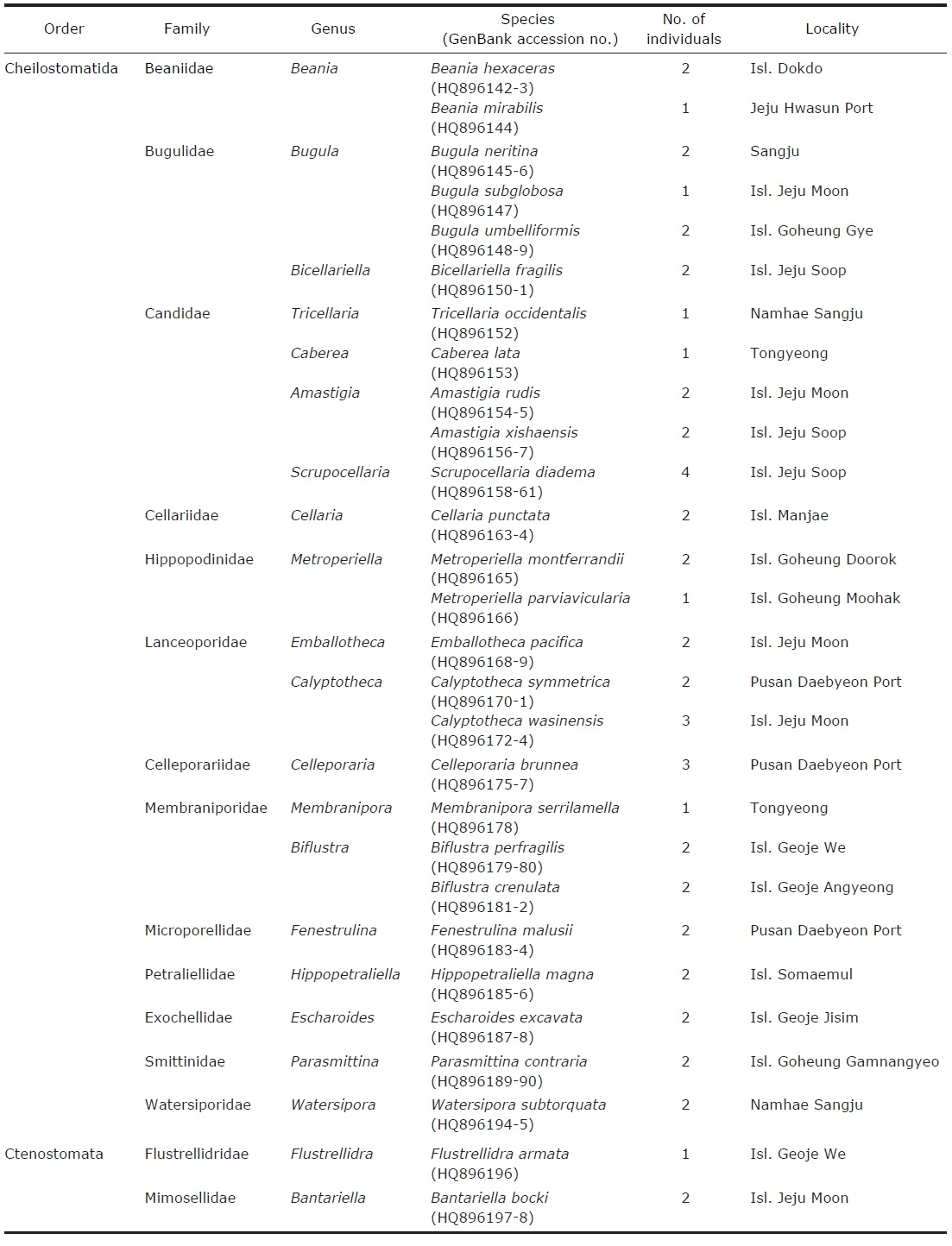
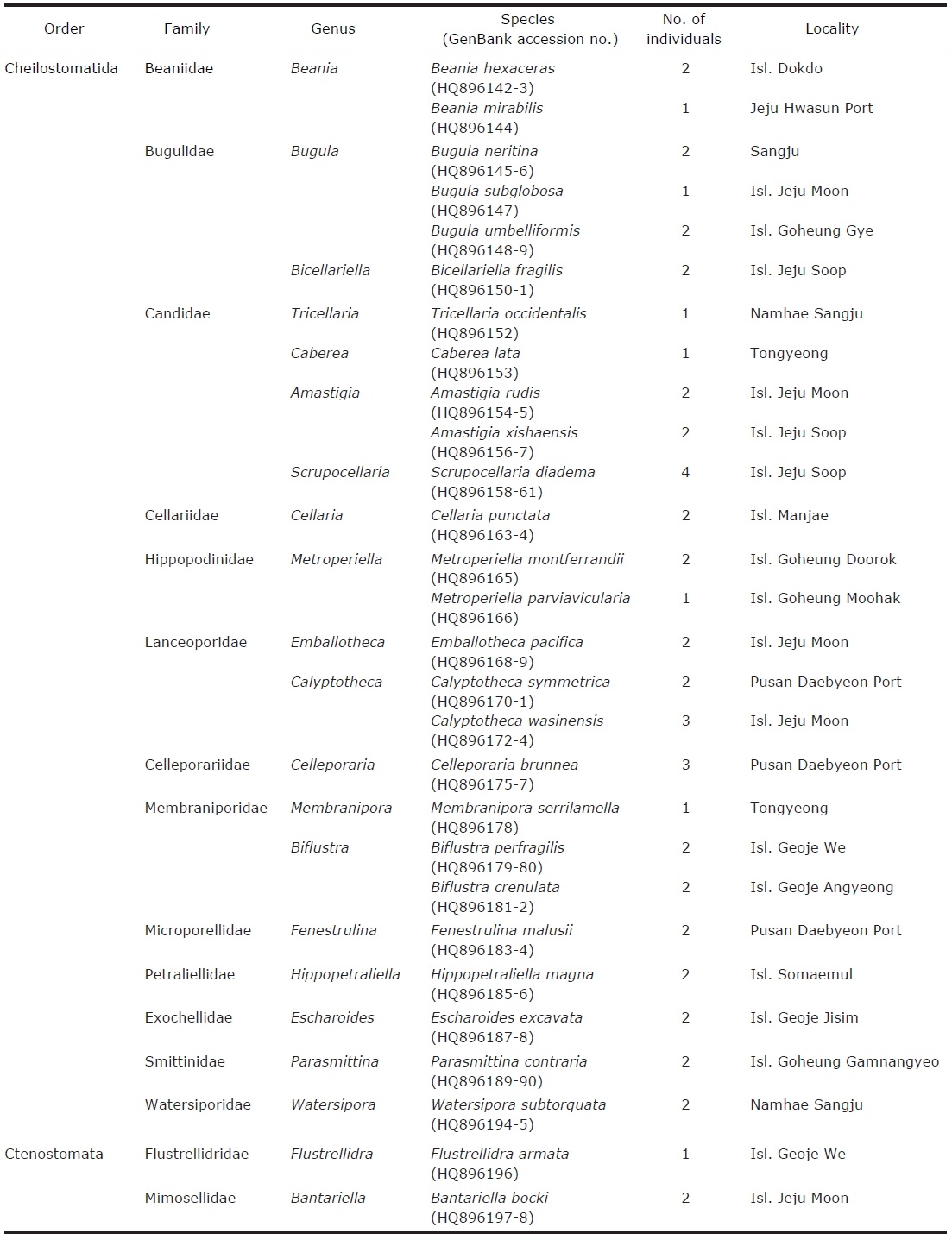
We attempted to construct COI barcodes based upon 53 individual samples from 29 species of Bryozoa collected from Korean seawater (Table 1). Genomic DNA was isolated from a small piece of each species using DNeasy Blood and Tissue Kits (QIAGEN Inc., USA) following the manufacturer’s protocol. A partial DNA fragment of mitochondrial COI was first amplified by PCR using the primers LCO1490 and HCO2198 (Folmer et al., 1994); however, the PCR products were not distinguishable on agarose gel. Thus, we designed a nested second PCR reaction using a degenerative BR_bugul_CO1_F (5′-CTG GDA TAR TTG GWA GAG G-3′) and BR_bugul_CO1_R (5′-GTR TTW AAA TTW
[Table 1.] Classification of Bryozoa (class Gymnolaemata) and GenBank accession numbers
Classification of Bryozoa (class Gymnolaemata) and GenBank accession numbers
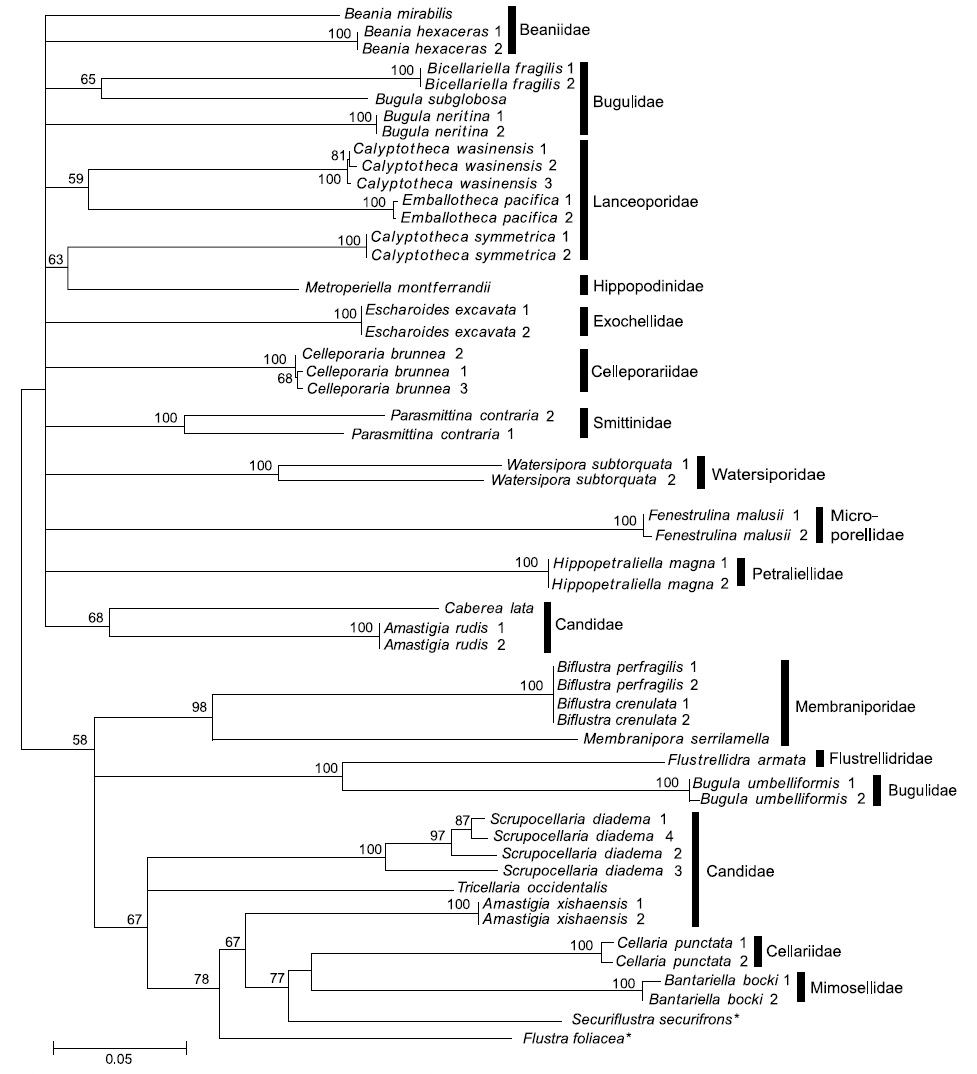
CGR TCK GTT A-3′) primer set specifically designed for the order Cheilostomatida, of which COI sequences were relatively abundant in GenBank. Fortunately, this new primer set was successful for cross-species amplification of COI from two species of the order Ctenostomata. The PCR products were purified with a LaboPassTM PCR purification kit (Cosmo Genetech Inc., Korea) and sequenced using an ABI 3730xl(Applied Biosystems Inc., USA). When we found mixed COI haplotypes due to a mixture of different individuals of conspecies,a typical problem of colony forming tissues in some specimens, we performed clone sequencing to phase out the ambiguous DNA sequence sites. Finally, pairwise sequence divergences and a neighbor-joining (NJ) tree were investigated with a Kimura-2-parameter (K2P) model using the computer software MEGA 4.0 (Kumar et al., 2004). GenBank accession numbers of the sequences that we deposited from this study and their taxonomical information are provided in Table 1.
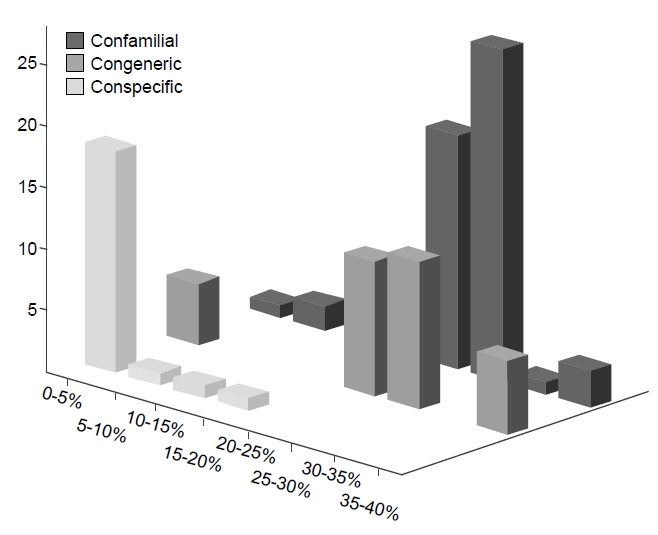
Our COI data was built from the largest taxon sampling to date, representing 29 species, 18 genera, 15 families, and 2 orders of Bryozoa. Assuming that the present COI data were obtained from the recognized species without error, the estimates of pairwise sequence divergence between species are as follows: 25% at a confamilial level, 23.3% at a congeneric taxonomic level, and 1.7% at a conspecific level (Fig.1 ).
Although Fig. 1 highlights increasing genetic divergence as taxonomic rank increases, the difference between generic and familial levels is still very small. Therefore, the main purpose of DNA barcoding for species identification has been fulfilled given the current taxon sampling with some exceptionally high divergent conspecific cases which strongly indicates the possibility of the presence of cryptic species in
Our study of bryozoans demonstrates the classic problems of species identification by molecular markers, which suffers from insufficient samplings, superficial knowledge of the range of molecular divergence, cryptic species, and phenotypic plasticity of recognized species (Vrijenhoek, 2009). In some cases, the COI marker did not provide resolution of species identification. As an extreme example, four sequences of
simple morphology, morphological species identification is difficult and, accordingly, multiple cryptic species could be involved. In contrast,
It should be noted that one of the most fundamental problems encountered with DNA barcoding of bryozoans was the lack of a universal COI primer set and insufficient reference sequences. During our study, we experienced frequent PCR failures with the universal Folmer primers (Folmer et al., 1994) for many bryozoans (not shown in the present results).This problem is not surprising if we take into account the fact that taxonomically broad bryozoans may have an enormous degree of COI divergence. In fact, the range of divergence and polymorphism of COI in bryozoans is yet unknown. The previously known COI sequences are limited to a very small portion of bryozoans, which actually impedes the design of universal primers. Hopefully, as COI data of bryozoans grow, development of universal primers should be possible. Such group specific oligonucleotide sequences might be desirable to minimize contamination due to nonbryozoan amplification, known as “the peril of universal primers” (Vrijenhoek, 2009). Another experimental difficulty was the frequent observation of mixed sequence reads from well-amplified PCR products, which indicates two possibilities:that is, either a mixture of variable haplotypes originating from a tissue specimen consisted of numerous tiny colonial individuals or a contamination of quite different species.Although the characteristic colonial structure of bryozoans imposes technical difficulties for extracting clean COI sequences from raw specimens. If we have suitable COI data from taxa across broad bryozoans taxa, the problem of mixed sequence reads will be more easily solved by comparing reference sequences well annotated with taxonomic information. In this regard, our current data, except for some suspicious cases,will add better discriminating power to the COI database in GenBank and BOLD for comparative analyses.
In summary, our DNA barcode study involving 29 bryozoan species is the first attempt of its kind in Korea. We found a high degree of COI sequence divergence among most species.The amount of differences was sufficient for species identification in most cases except for two species. We therefore believe that COI can be served as a standard molecular maker for DNA barcoding of bryozoans.