During vitellogenesis in oviparous animals,nutrient-rich egg yolk composed mostly of lipids and proteins accumulates. Vitellin (Vn) is the major protein component in the yolk, and it is synthesized as a precursor protein, vitellogenin (Vg) (Gerber-Huber et al., 1987; Chen et al., 1997). Vg is one of the most important proteins during oocyte maturation,and numerous studies have examined its characteristics and roles during the reproductive cycle.Furthermore, the Vg genes have been studied as representative markers induced by endocrinedisrupting chemicals (EDCs).
Due to its possible industrial applications, such as in seed production, the Vgs from commercially important decapod crustaceans have been studied. To our knowledge, 19 crustacean Vg cDNAs have been isolated and characterized. Most are from the commercially important penaeid shrimps in the suborder of Dendrobranchiata, including Marsupenaeus japonicus (Tsutsui et al., 2000), Penaeus monodon (Tiu et al., 2006), Litopenaeus vannamei(Raviv et al., 2006), Penaeus semisulcatus (Avarre et al., 2003), Fenneropenaeus chinensis (Xie et al.,2009), Fenneropenaeus merguiensis (Phiriyangkul and Utarabhand, 2006), and Pandalopsis japonica(Jeon et al., 2010). Others are from brachyuran and astacidean species in the suborder Pleocyemata,including Macrobrachium rosenbergii (Okuno et al.,2002), Pandalus hypsinotus (Tsutsui et al., 2004),Portunus trituberculatus (Yang et al., 2005), Cherax quadricarinatus (Abdu et al., 2002), Homarus americanus (Tiu et al., 2009), Upogebia major (Kang et al., 2008), Charybdis feriatus (Mak et al., 2005),Portunus trituberculatus (Yang et al., 2005), and Callinectes sapidus (Zmora et al., 2007).
Compared with the Vg cDNA sequences, only a few genomic analysis of Vg genes from decapod crustaceans have been conducted, mainly due to the large gene size (>10 kb), which makes it difficult to isolate full-length Vg genes. However, recent advances in molecular techniques and the development of genomic analysis tools enabled us to obtain Vg cDNA sequences with less effort. The full-length crustacean Vg genes have been determined only in three species: Metapenaeus ensis (Tsang et al., 2003;Kung et al., 2004), P. monodon (Tiu et al., 2006), and H. americanus (Tiu et al., 2009).
The accumulated information on Vg in decapod crustaceans raised two major questions. First, how many copies of Vg genes do these animals possess?Until now, the full length of two distinct Vg genes has been determined only in M. ensis (Tsang et al., 2003;Kung et al., 2004). However, several studies support the idea that decapods possess multiple Vg genes(Mak et al., 2005; Tiu et al., 2009). Recently, we first isolated two distinct copies of Vg genes from a species belonging to the suborder Pleocyemata: the Pandalus shrimp, P. japonica (Jeon et al., 2010). It is necessary to analyze all known crustacean Vgs to determine structural and biological differences.Second, where is the major site of Vg production?Despite contradictory results, it is generally accepted that the major production site for Vg is the hepatopancreas, and the synthesized Vg is understood to be processed into Vn, circulated, and transported into the oocyte via receptor-mediated endocytosis.However, several studies have shown that the ovary is another source of Vg production, and the similarities and differences in the hepatopancreatic and ovarian expression of Vg need to be investigated.
Pandalopsis japonica is a commercially important species in the coastal regions of eastern Asia,including Korea and Japan. Recently, its production has been declining due to a combination of factors,including climate change, overfishing, and pollution.To restore its population, it is important to develop a seed-production technique. However, little is known about the endocrinology and molecular mechanisms of crustacean reproduction. Previously, we isolated two distinct Vg cDNAs, Pj-Vg1 and Pj-Vg2, from P.japonica and found that their downregulated by the major component of insecticides,endosulfan (Jeon et al., 2010). In this study,we isolated and analyzed the two full-length Pj-Vg1 and Pj-Vg2 genes. To estimate their roles in crustacean reproduction, we also analyzed the relationship of the expression of the two Pj-Vgs to the reproductive stage and production sites.
Live female P. japonica were purchased from a local seafood market. The live shrimp were dissected quickly after sacrifice. Isolated tissues were deepfrozen by immersing them in liquid nitrogen. The frozen tissues were stored at -70°C until use. The individual gonad-somatic index (GSI=gonad weight/body weight) was also determined for further study.
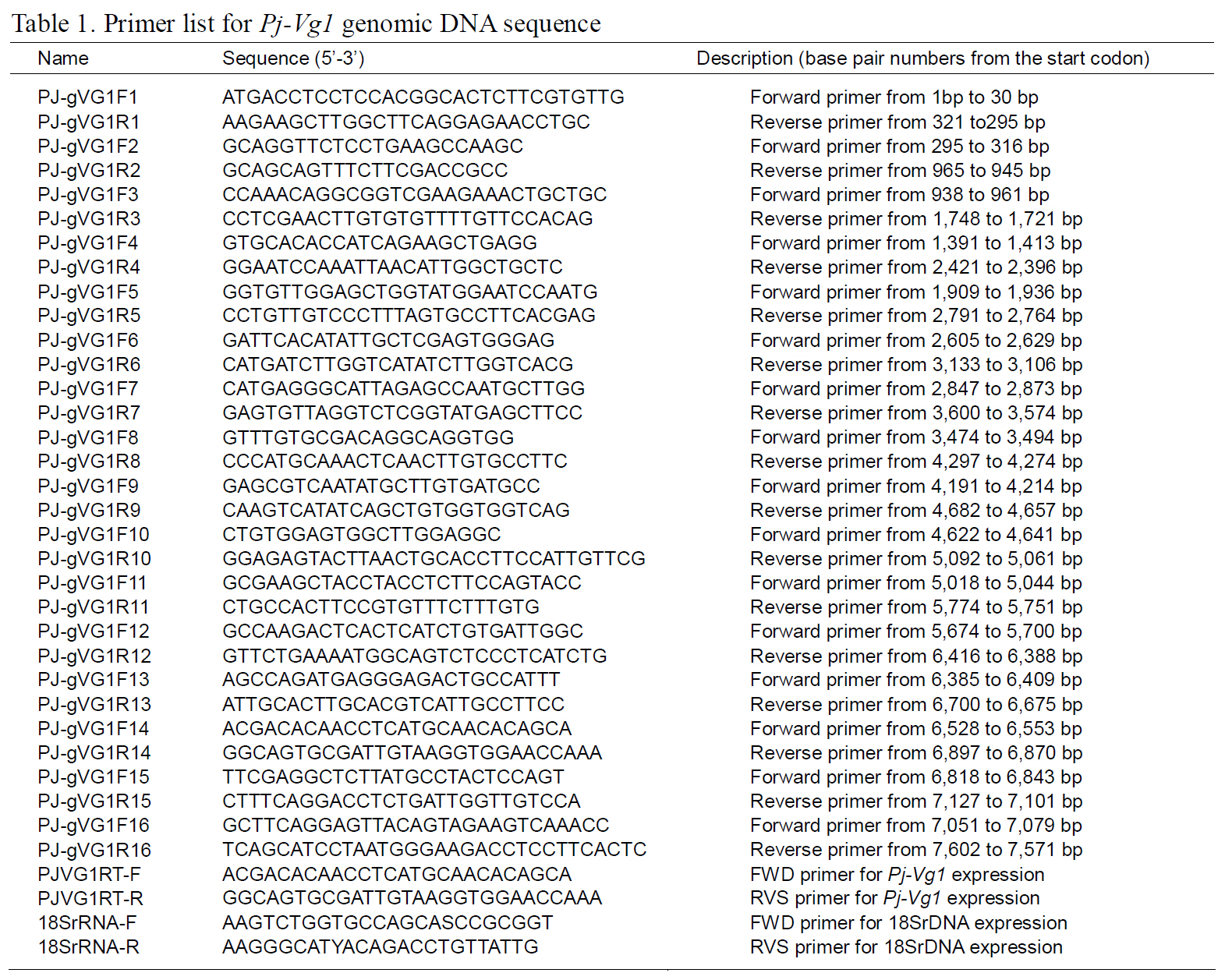
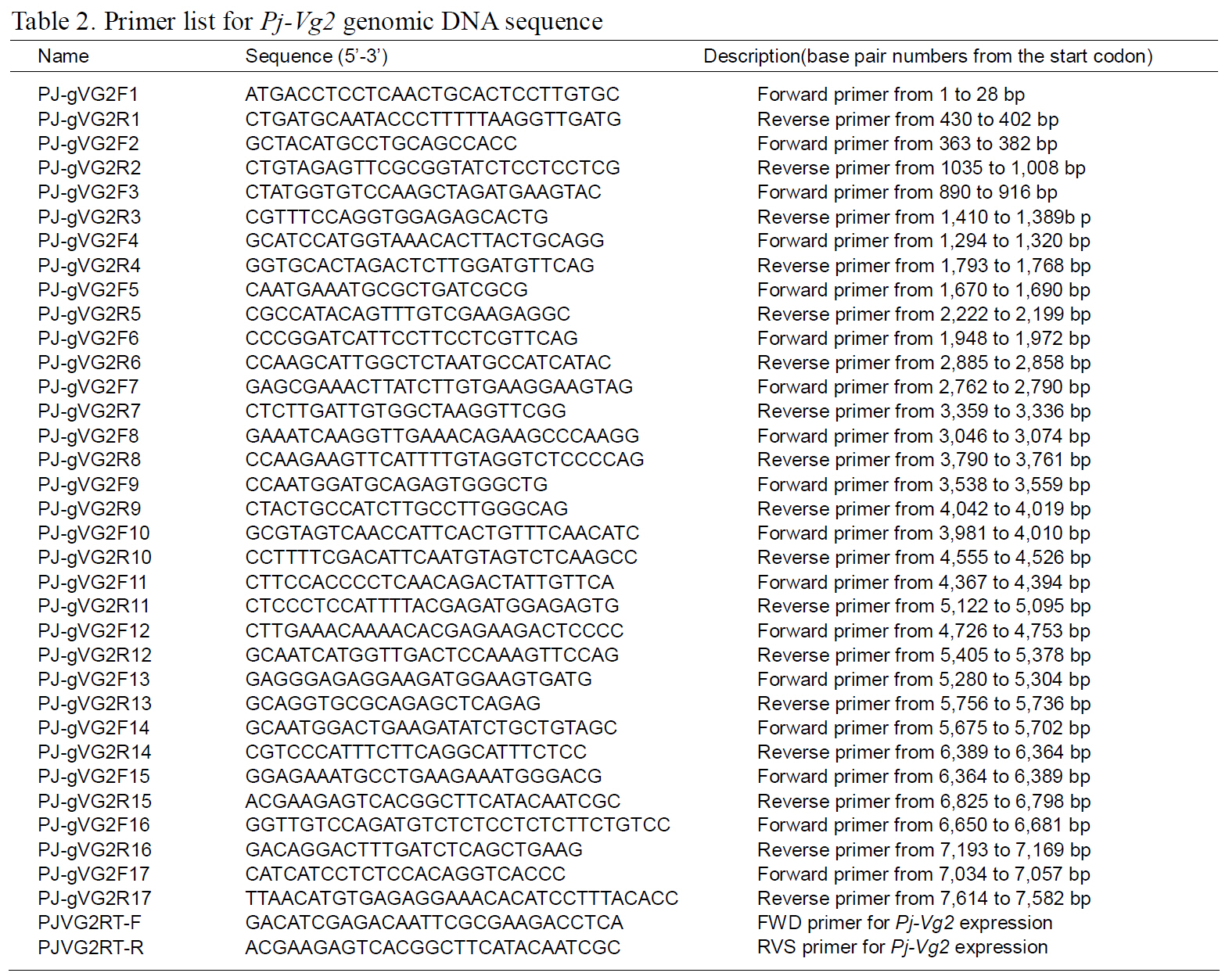
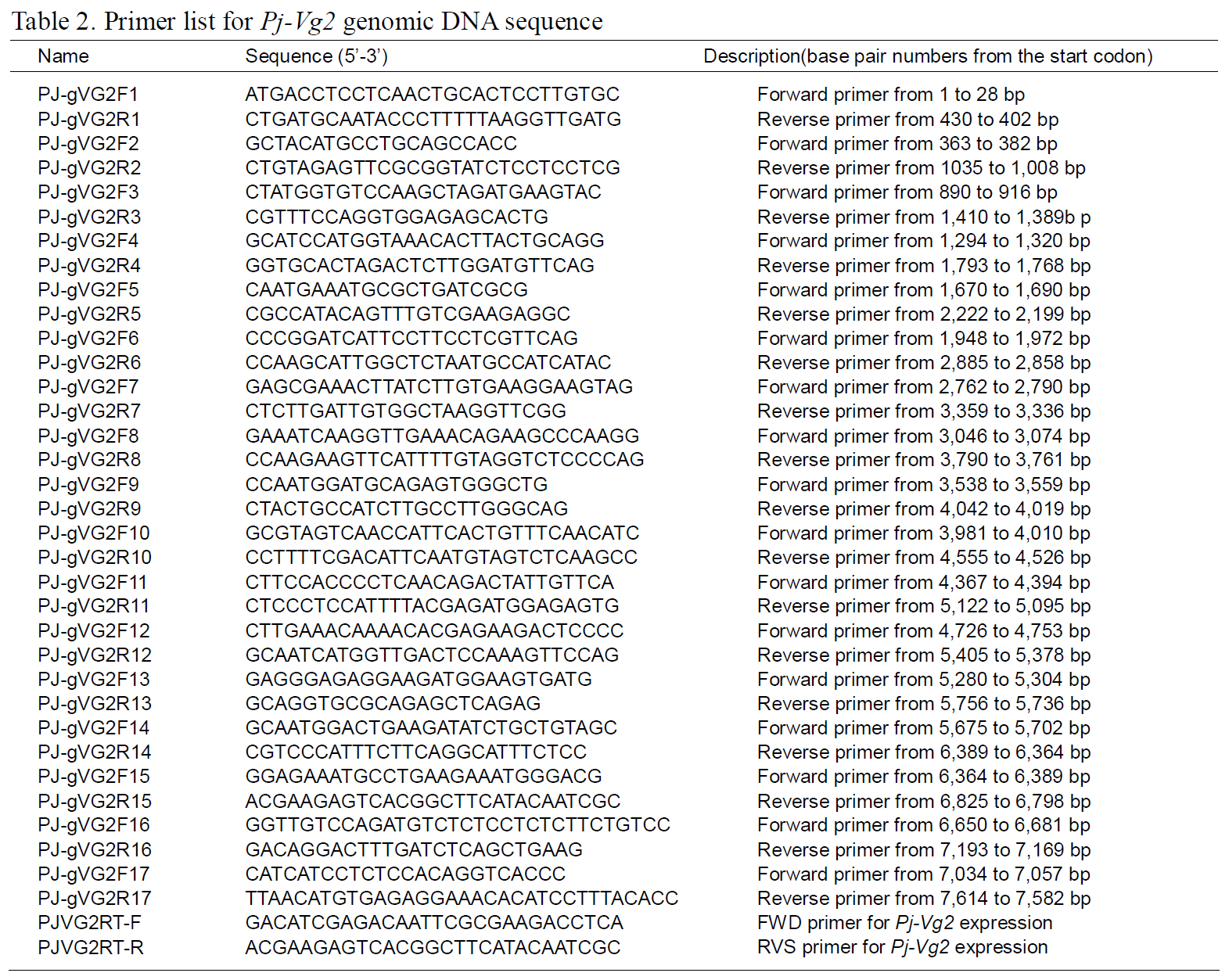
The structures of the two Pj-Vg genes were determined using a PCR-based strategy. First,genomic DNA was purified from muscle tissue using an AccuPrep® Genomic DNA Extraction Kit (Bioneer,Korea). Then, 200 ng of genomic DNA were used for each PCR reaction. Given the expected large size of each Pj-Vg gene, several primers were designed to amplify products smaller than 2 kb to avoid landing on an exon-intron boundary (Table 1). Some PCR products that were too large to be cloned into Tvector were digested with Sau3AI, mixed with Taq polymerase and dNTPs, and incubated at 72°C for 10 min. The smaller fragments were ligated into pGEM T-easy vector (Promega, USA). The full lengths of the Pj-Vg1 and two genomic sequences were determined by combining the fragment sequences.The resulting contig sequences were then confirmed by PCR using two primer pairs (PJ-gVG1F1 and PJgVG1R16;PJ-gVG2F1 and PJ-gVG2R17), which land on both the 5' and 3' ends (Table. 1 and 2).Exons and introns were determined by comparing the cDNA sequences with the obtained genomic DNA sequences.
To understand the expression of the two Pj-Vgs,quantitative PCR analysis was carried out. Fourteen females with various GSI indices were sacrificed to obtain the ovary and hepatopancreas. Total RNA was purified using TRIzol reagent according to the
manufacturer’s instructions (Invitrogen, USA),quantified using the absorbance at 260-nm(NanoDrop Technologies, USA), and stored at -70°C.Before cDNA synthesis, the total RNA was pretreated with DNase I (Takara, Japan) for 20 min at 37°C to remove any genomic DNA contamination. Then, the cDNA was synthesized in a reaction (12-μL)containing 1 μg of total RNA, 1 μL of 20 μM random hexamer, and 4 μL of dNTPs (2.5 mM each). The mixture was heated to 70°C for 5 min and then chilled on ice for 2 min. First-strand buffer (5×, 4 μL),2 μL DTT (0.1 M), and 1 μL RNaseOUT (Invitrogen,USA) were added, and the reactant was incubated at 37°C for 2-min. Finally, 1-μL MMLV reverse transcriptase (Invitrogen, USA) was added, and the mixture was incubated at 37°C for 50 min. The synthesized cDNA was quantified and stored at -20°C before being used for PCR.
Quantitative PCR was carried out using the DNA Engine Opticon 3 Real-Time PCR Detection System(Bio-Rad, USA) and SYBR Green premix Ex Taq II(Takara Bio, Japan) to measure the mRNA transcripts.Sequence-specific primers for each target gene were designed using PrimerQuest (http://www.idtdna.com).The reaction mixture (20 μL) contained 1 μL cDNA(100-ng), 2-μL 4-μM sequence-specific primers,which are shown in Table. 1 and 2 (PJVG1 RT-F and PJVG1 RT-R for Pj-Vg1, PJVG2 RT-F and PJVG2 RT-R for Pj-Vg2), and autoclaved deionized water to a volume of 20 μL. The PCR conditions were 1 min at 94°C followed by 40 cycles at 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s. Standard curves were constructed to confirm the efficiency of the primers and to quantify copy numbers, as described previously (Kim et al., 2005). The Pj-Vg copy numbers were normalized to the 18S rRNA copy number using to the following equation: (actual copy numbers of Pj-Vg1 or Pj-Vg2 / actual copy number of
18S rRNA)×10,000.
Multiple amino-acid sequence analysis was performed using ClustalW software (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and presented using GeneDoc software (http://www.nrbsc.org/gfx/genedoc/index.html). The Pj-Vg analyzed statistically by the independent t-test using the Statistical Package for the Social Sciences (SPSS)(version 10.1.3). The results were considered significant at P<0.05.
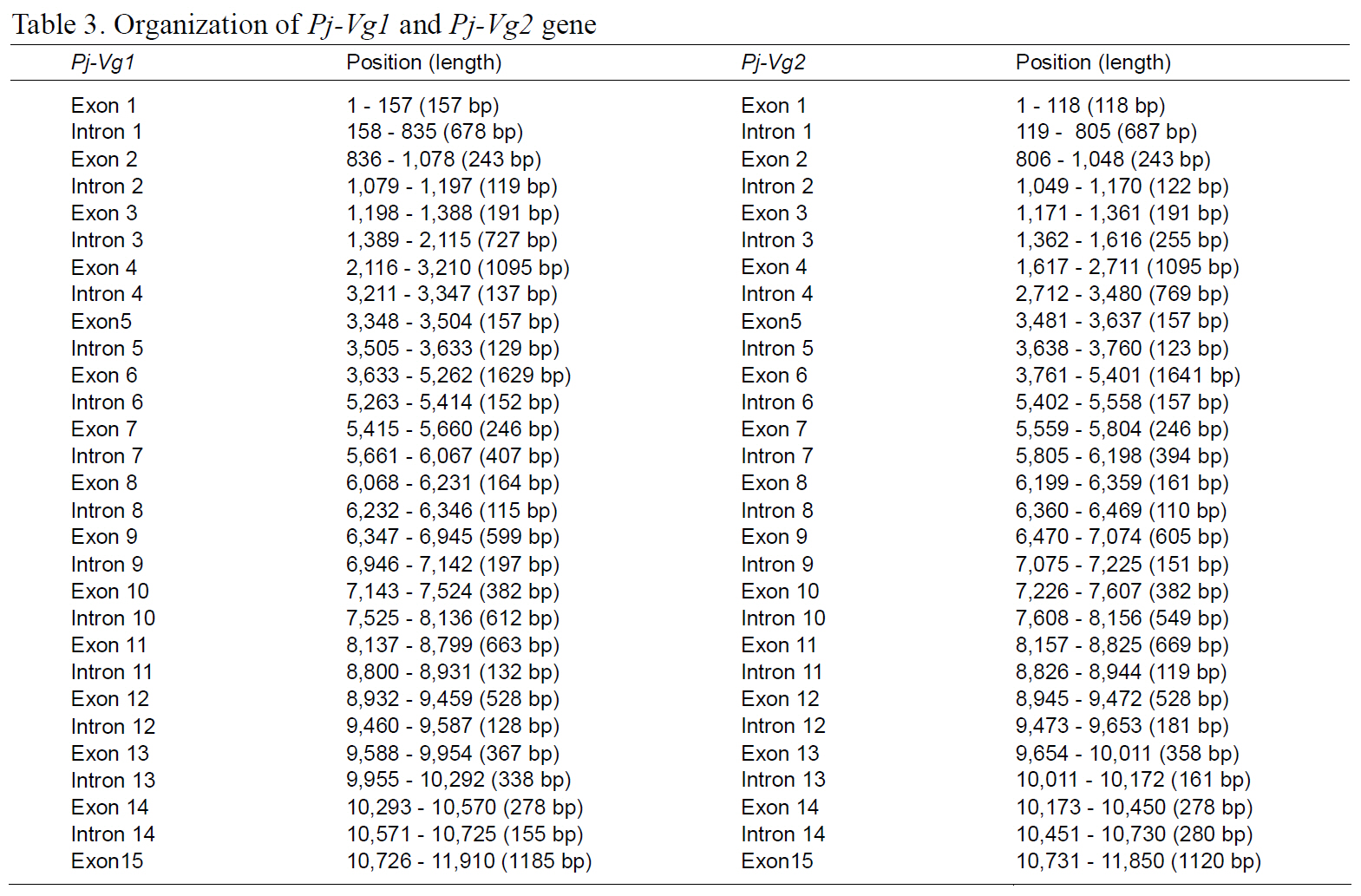
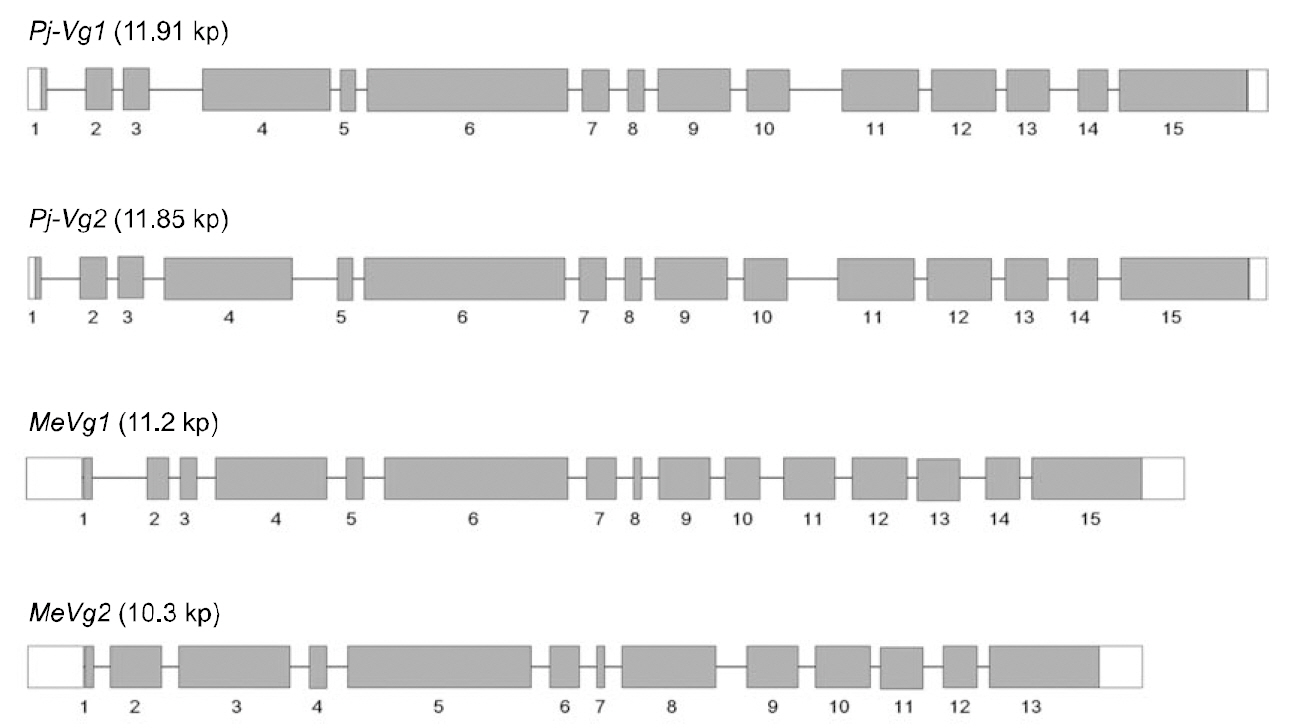
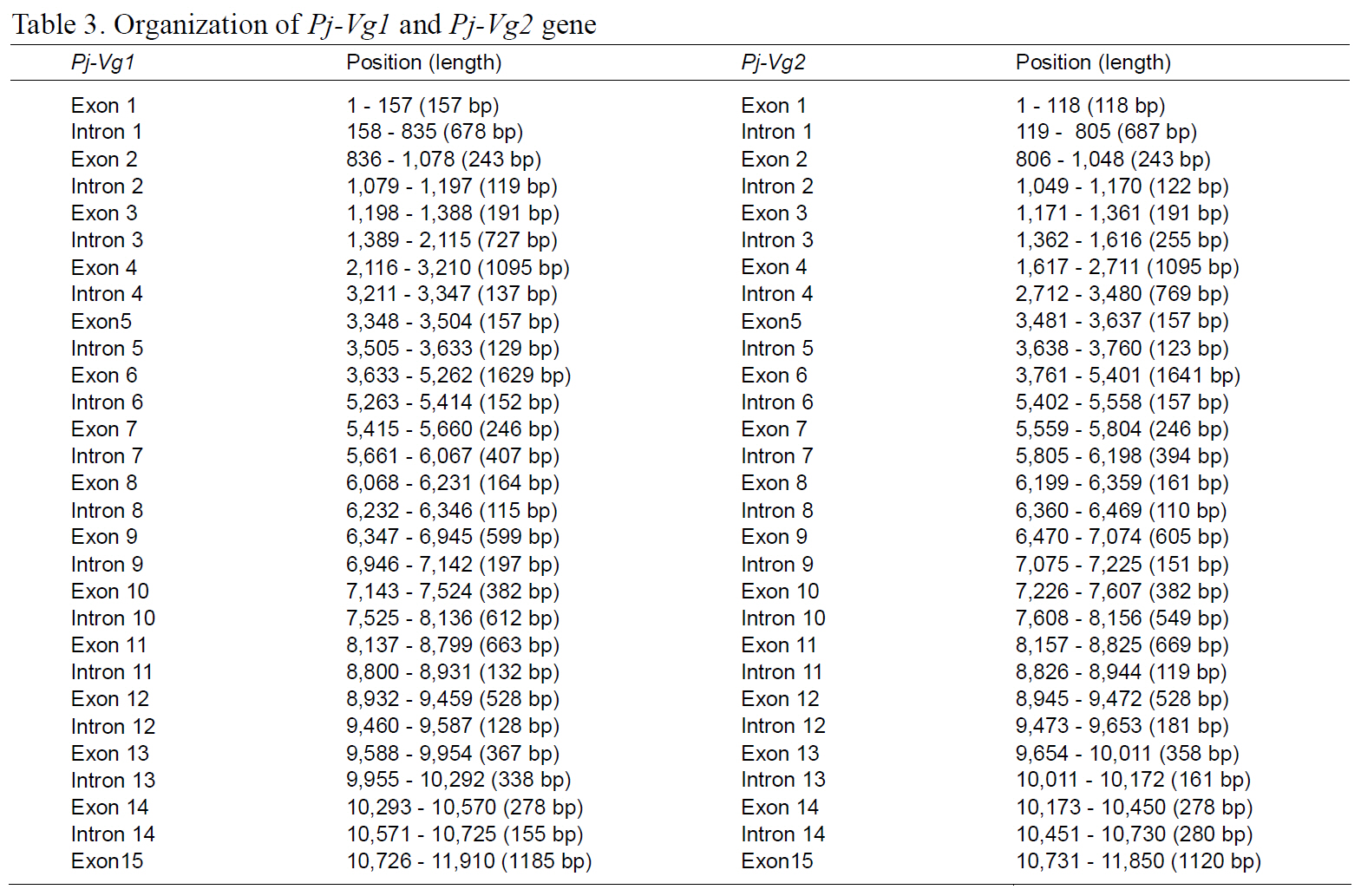
The full-length Pj-Vg1 and Pj-Vg2genes were obtained using a PCR-based strategy. The full-sized genomic DNA sequences assembled from the PCR products were confirmed by determining the nucleotide sequence of the large PCR products (-11 kb) generated using sequence-specific 5'- and 3'-end primers (Table. 1 and 2). The Pj-Vg1 and -Vg2 genes were 11,910 and 11,850 bp long, respectively, which is similar to the Vg genes from other decapod crustaceans (Fig. 1). To our knowledge, only three genomic structures of decapod Vg genes are known: P. monodon (Tiu et al., 2006), M. ensis (Tsang et al.,2003; Kung et al., 2004), and H. americanus (Tiu et al., 2009). Of these, the structures of two copies of the Vg gene were determined only in M. ensis, and only single gene structures were determined in the other two species. Our study is the first report on the genomic structure of two copies of Vg genes in the suborder Pleocyemata.
Structural characteristics of the Pj-Vg genes were analyzed, revealing that all of the exon-intron boundaries of the Pj-Vgs followed the general
spliceosome sequence rule (GT-AG, Shapiro and Senapathy, 1987). The overall sizes of the exons and introns were also analyzed (Table 3). The sizes of each intron in the Pj-Vgs were similar, except for the third and fourth introns (Fig. 1). The average size of the introns in Pj-Vg1 and Pj-Vg2 was 287 and 289 bp,respectively. These relatively small intron sizes appear to be common characteristics of decapod crustacean Vg genes, in which the average intron size ranges from 100 to 300 bp (Kung et al., 2004; Tiu. et al., 2006). Both Pj-Vg1 and Pj-Vg2 have 15 exons and 14 introns (Fig. 1). All reported Vg genes from the three other decapod crustaceans have the same numbers of exons and introns as the Pj-Vg genes,except MeVg2, which has 13 exons as a result of the fusion of exons 2 and 3 and of exons 9 and 10 (Kung et al., 2004). The exon?intron boundaries of Pj-Vg1,Pj-Vg2, and MeVg1were also compared (Fig. 2).Interestingly, all of the splicing sites were conserved in all three Vg genes. This suggests that the three Vg genes are orthologs and likely have evolved from the same ancestral gene. MeVg2 and the other genomic sequences were not used in the analysis because they are not yet publicly available.
In this study, we isolated and characterized two distinct Vg genes for the first time from a species in the suborder Pleocyemata: P. japonica. Multiple copies of Vg genes have also been identified in M.ensis in the suborder Dendrobranchiata. Although the full-length gene has not been isolated, data suggest that multiple copies of Vg genes are not limited to those species. In the infraorder Astacidea, several fragment Vg sequences subcloned from H.americanus differed slightly, supporting the existence of multiple Vg genes (Tiu et al., 2009). In brachyuran species, several sequences had distinct sequences,suggesting that there are two copies of the Vg genes(Mak et al., 2005). Two Vg cDNA sequences obtained from the penaeid shrimp M. japonicus differed at 47 nucleotide positions. The author proposed that this arose from individual variation and provided no experimental evidence of two distinct genes (Tsutsui et al., 2005). However, our finding of two copies of Vg genes in the suborder Pleocyemata leads us to postulate that the presence of two copies of the Vg gene is a general characteristic of decapod crustaceans.
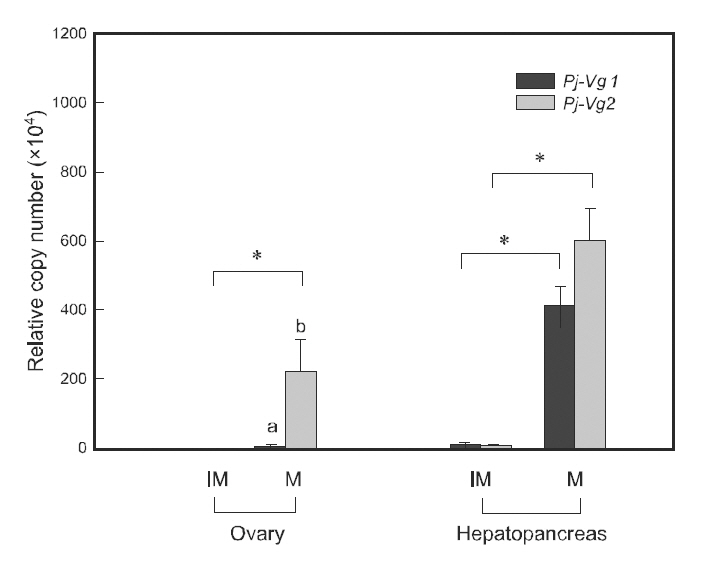
Quantitative PCR was used to examine the relationship between the expression of Pj-Vgs and the reproductive cycle. Individual maturation was determined using the GSI, as described in Kim et al.(2006). The expression of both Pj-Vg1 and Pj-Vg2 transcripts was at a basal level in the hepatopancreas and ovary of immature individuals (GSI 0.2-1.3).Although their expression levels were extremely low in immature animals, both shrimp Vg transcript copy numbers were significantly higher in the hepatopancreas than in the ovary (200-fold for Pj-Vg1 and 50-fold for Pj-Vg2). The expression of both Pj-Vgs in mature individuals (GSI 2.1-9.8) was also significantly higher in the hepatopancreas than in the ovary. The expression of Pj-Vg1 and Pj-Vg2 transcripts was 97- and 2.7-fold higher, respectively,in the hepatopancreas than in the ovary, suggesting that the major production site for both Pj-Vgs is the hepatopancreas regardless of the stage in the reproductive cycle. The high copy numbers of Pj-Vg2 in the ovaries from mature animals were noteworthy.The induction of transcripts of both Pj-Vg1 (34.6-fold) and Pj-Vg2(104.1-fold) was greater in the hepatopancreas from mature individuals compared with immature animals.
It is generally accepted that the expression of Vg genes is upregulated in the hepatopancreas during the mature stages. The induction of Vg genes in the ovary has also been detected in various species, including P.monodon (Tiu et al., 2006), M. ensis (Tsang et al.,2003), Callinectes sapidus (Zmora et al., 2007), and Upogebia major (Kang et al., 2008). In our study,there was a 1972-fold induction of Pj-Vg2transcripts in the mature ovary, whereas no significant increase in Pj-Vg1 was seen. To our knowledge, this is to first report on the different expression patterns of two Vg genes. The existence of two mechanisms regulating Vg genes during the reproductive cycle is of interest.Further study is needed to elucidate the cis- and trans-elements that are responsible for regulating this and their biological roles. There was no statistical difference between Pj-Vg1 and Pj-Vg2 in the mature hepatopancreas.
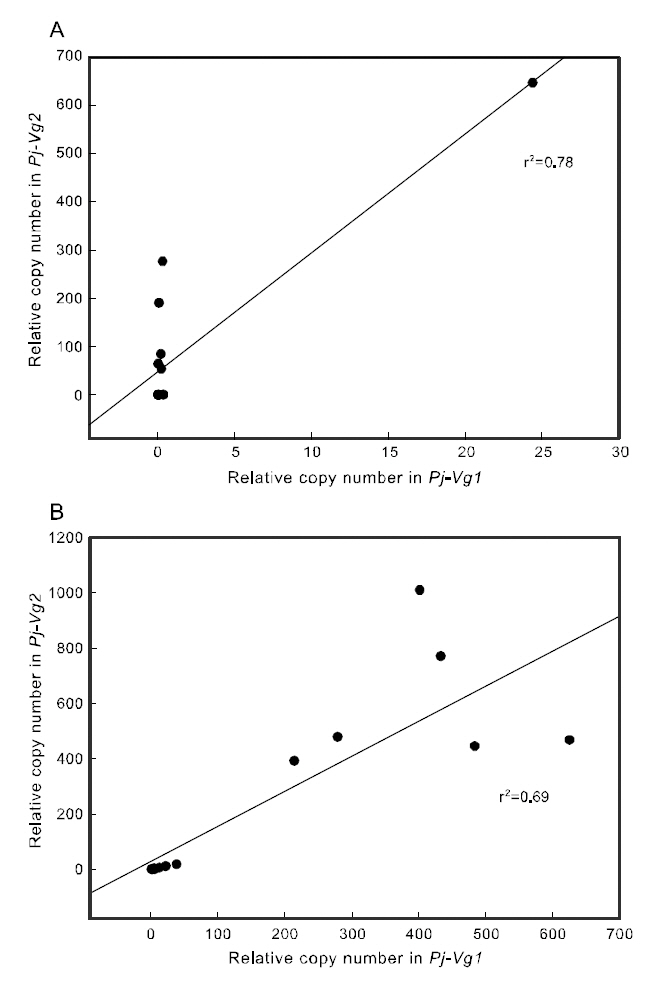
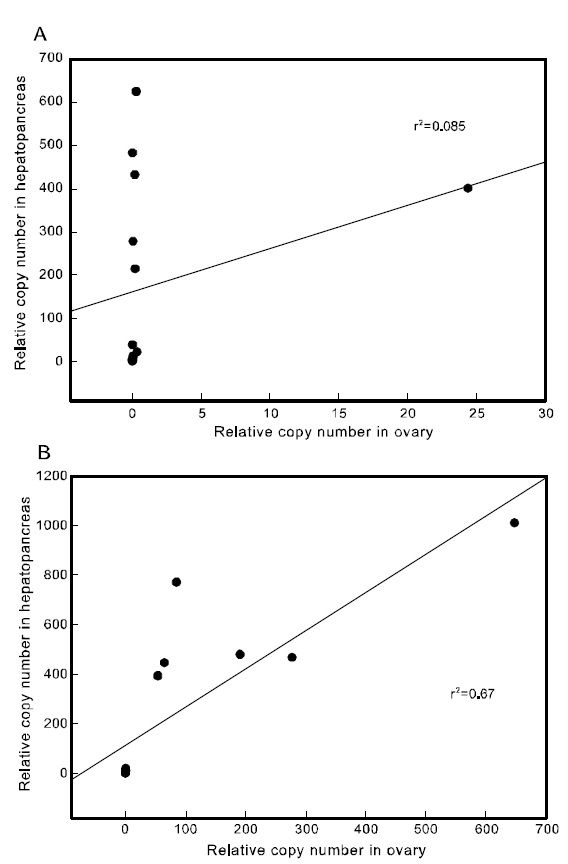
To examine the correlation between the expression of the two Pj-Vgs within the same individual, a simple regression analysis was carried out. The expression of Pj-Vg1 and that of Pj-Vg2 were strongly correlated in both the gonad and hepatopancreas(Fig. 4). The relative copy numbers of Pj-Vg2 in the hepatopancreas were also correlated with those in the ovary, suggesting that the dual expression of Pj-Vg2 in the hepatopancreas and ovary is under
the same regulatory mechanism during the reproductive cycle (Fig. 5). As no Pj-Vg1 was expressed in the immature hepatopancreas, we could not examine this relationship for Pj-Vg1. We conclude that the expression of Pj-Vg1 and that of Pj-Vg2were strongly correlated in the hepatopancreas, and the induction of Pj-Vg2in both the hepato-pancreas and ovary was synchronized. The meaning of the absence of Pj-Vg1 in the ovary is not clear, but our findings constitute important data for under-standing the roles of multiple vitellogenin genes in the reproductive mechanism of decapod crustaceans.