Most species of the family Gelidiaceae are familiar to molecular biologists because they produce commercially important agar for laboratory gels and bacterial growth media; they are also a food source. The Gelidiaceae is the largest family in the order and includes nine genera,viz. Acanthopeltis Okamura in Yatabe, Beckerella Kylin,Capreolia Guiry & Womersley, Gelidium Lamouroux,Onikusa Akatsuka, Porphyroglossum Kutzing, Ptilophora Kutzing, Suhria J. Agardh ex Endlicher, and Yatabella Okamura. The family is characterized by brush rhizoids and by endofibers in cortical and medullary layers (Perrone et al.2006).
Gelidium is the largest genus within the family, comprising about 110 species widely distributed throughout tropical and temperate regions. Most members of the genus have large erect thalli with slightly protruding apical cells, tetrasporangia that are usually scattered on terminal branchlets, bilocular cystocarps with single, terminal carposporangia, and isomorphic life histories. Three large genera, Gelidium, Pterocladiella, and Ptilophora, are clearly established (e.g., Freshwater et al. 1995, Kim et al. in press), whereas smaller genera with one or few species are under taxonomic reconsideration. For example,Shimada et al. (1999) compared nuclear ribosomal cistron small subunit (SSU) and internal transcribed spacer (ITS) genes and plastid rbcL genes between Acanthopeltis and Yatabella and concluded that these entities are congeneric. Tronchin et al. (2002) reduced Onikusa and Suhria to synonymy under Gelidium because rbcL tree analysis nested both within Gelidium; Onikusa and Suhria have female reproductive and cystocarp systems very similar to those of Gelidium. Beckerella has also been treated as a congener of Ptilophora based on analyses of the large subunit (LSU) of the nuclear cistron and rbcL (Tronchin et al. 2003). Kim et al. (in press) demonstrated through three-gene sequence analysis that Acanthopeltis is conspecific with Gelidium. In contrast, Capreolia is well supported by molecular data (Nelson et al. 2006). Porphyroglossum is the last genus within the family that has yet to be analyzed by molecular markers.
Porphyroglossum zollingeri Kutzing is a candidate species for mass culture in Indonesia. Porphyroglossum zollingeri, the only species in the genus, was described from specimens collected in Java, Indonesia (Kutzing 1847). Later, Schmitz (1894) added a second species, P. japonicum (= Suhria japonica Harvey). However, Akatsuka (1986) moved this second species to his new genus Onikusa, leaving a single species in Porphyroglossum. Porphyroglossum is distinguished by broad axes densely trimmed with slender lanceolate pinnules along the midline; it is attached to substrata by peg-like haptera (Kutzing 1847, Kylin 1956, Hatta and Prud’homme van Reine 1991). The genus is currently recognized as an independent entity in the list of benthic marine algae in the Indian Ocean (Silva et al. 1996) and in AlgaeBase (Guiry and Guiry 2011). However, with their detailed observations of morphology, Hatta and Prud’homme van Reine (1991) cast doubt on the taxonomic validity of Porphyroglossum.
Molecular markers have helped to correctly assign taxonomically confused taxa within the order Gelidiales. Earlier studies on gelidoid algae used only nuclear ribosomal cistron (small and large subunits and their spacer) and plastid rbcL genes for identification and phylogenet-
ic analysis (Freshwater and Rueness 1994, Freshwater et al. 1995, Millar and Freshwater 2005, Nelson et al. 2006). Recently, Kim et al. (in press) found that mitochondrial cox1 genes are also suitable for classifying these commercially important algae. In this study, we analyzed rbcL and cox1 from 16 specimens of P. zollingeri from Indonesia and reconstructed phylogenetic trees of sequence data for the two genes to determine whether Porphyroglossum is supported by molecular data.
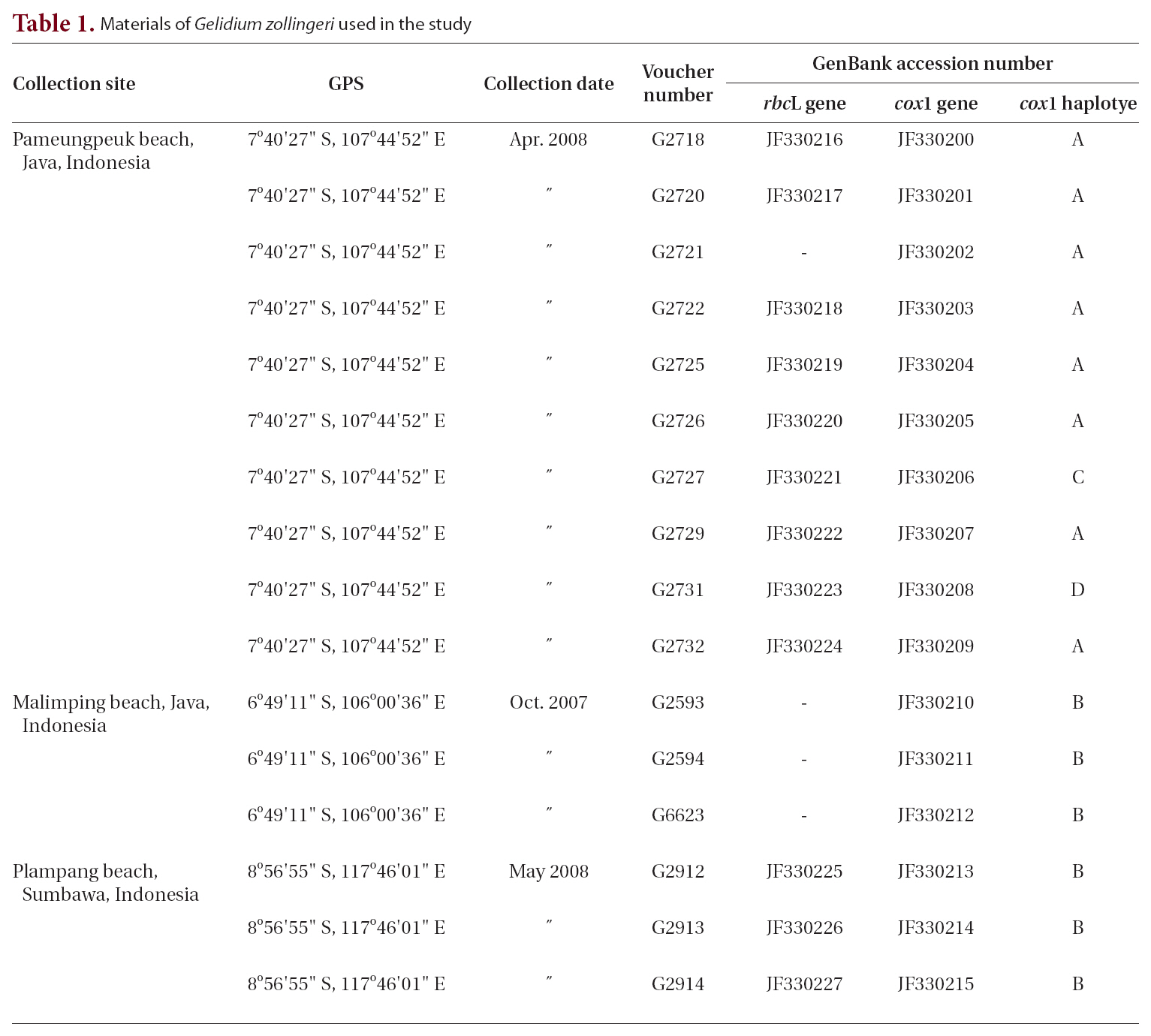
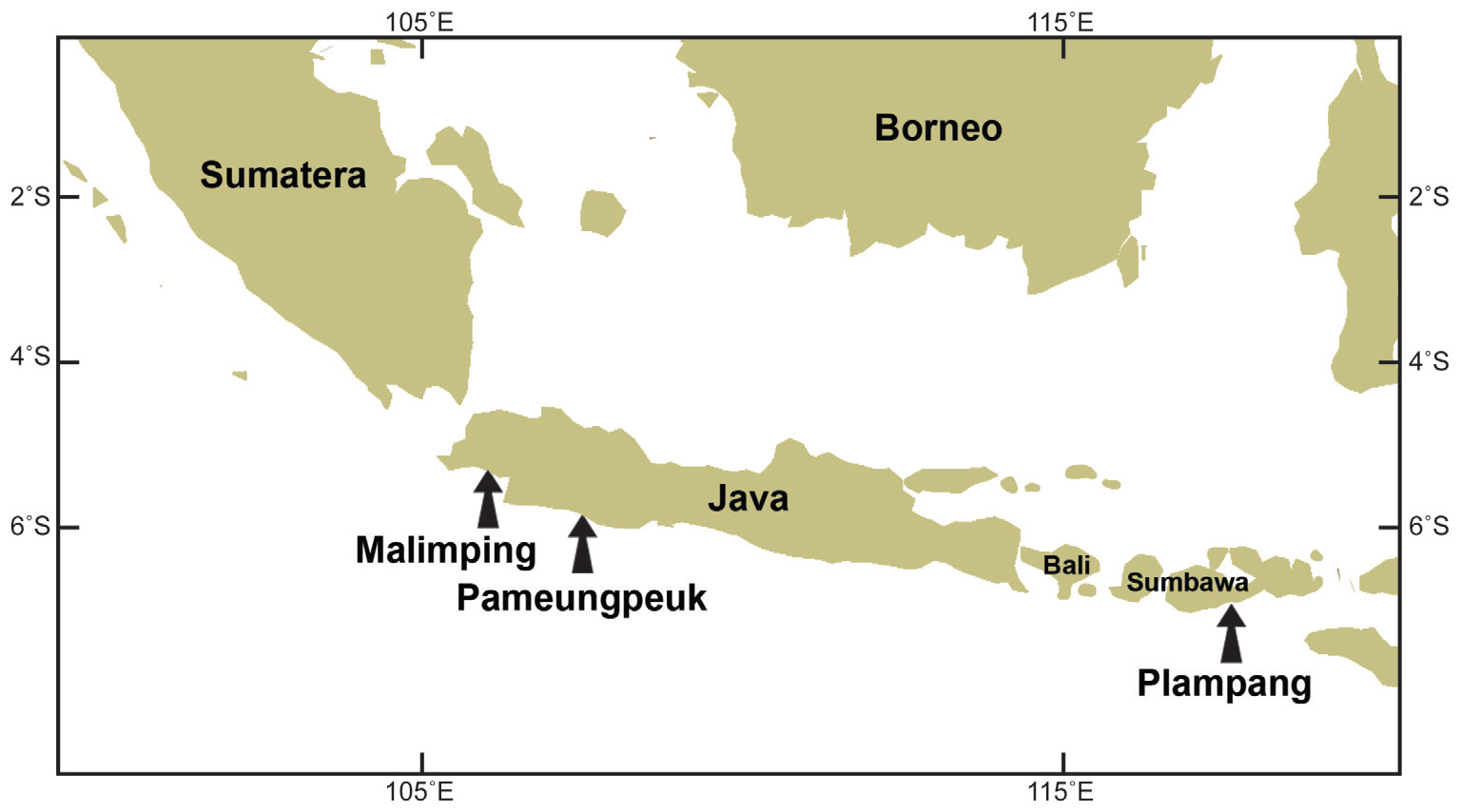
Field collections of Porphyroglossum were made at three locations in Indonesia (Table 1, Fig. 1). Tissues were sectioned with a freezing microtome (FX-802A; Coper Electronics Co., Ltd., Kanagawa, Japan) and sections were stained with 1% aqueous aniline blue. Photographs were taken with an FX-35DX camera (Nikon, Tokyo, Japan) attached to a Vanox AHBT3 microscope (Olympus, Tokyo, Japan). Voucher specimens are housed at the herbarium of Chungnam National University, Daejeon, Korea (CNUK).
Sixteen specimens were available for extraction of DNA (Table 1). DNA extraction, PCR amplification, and sequencing are described in Geraldino et al. (2010). Primer pairs for amplification and sequencing of each gene were as follows: for rbcL, F7-R753 and F645-RrbcS start (Freshwater and Rueness 1994, Lin et al. 2001, Gavio and Fredericq 2002); and for cox1, cox143F-cox11549R (Geraldino et al. 2006) and C622F-C880R (Yang et al. 2008).
Sixty-eight rbcL sequences (12 new and 56 published) from P. zollingeri, Acanthopeltis, and Gelidium including outgroups were collated using the Se-Al version 2.0 a11 software (Rambaut 1996) and aligned visually. Outgroup taxa included Pterocladia, Pterocladiella, and Ptilophora. Phylogenies were rooted with the distantly related genus Gelidiella Feldmann and G. Hamel (Freshwater et al. 1995, Kim et al. in press).
Maximum likelihood (ML) phylogenetic analysis of rbcL was performed using only the GTR + Γ + I model implemented in RAxML software (Stamatakis 2006). We used 100 independent tree inferences with the “number of run” option, with default optimized SPR rearrangement and 25 distinct rate categories to identify the best tree. Statistical support for each branch was obtained from 1,000 bootstrap replications using the same substitution model and RAxML program settings.
Maximum parsimony (MP) tree was constructed for each data set with PAUP* version 4.0b.10 software (Swofford 2002) using a heuristic search algorithm with the
Porphyfollowing settings: 1,000 random sequence additions, tree bisection-reconnection (TBR) branch swapping, MulTrees, all characters unordered and unweighted, and branches with a maximum length of zero collapsed to yield polytomies. Bootstrap values for the resulting nodes were assessed using 1,000 bootstrapping replicates with ten random sequence additions, TBR, and MulTrees.
A statistical parsimony network of cox1 haplotypes was created using TCS version 1.21 software (Clement et al. 2000). Haplotype and nucleotide diversity measurements were performed using DNAsp software (Rozas and Rozas 1999).
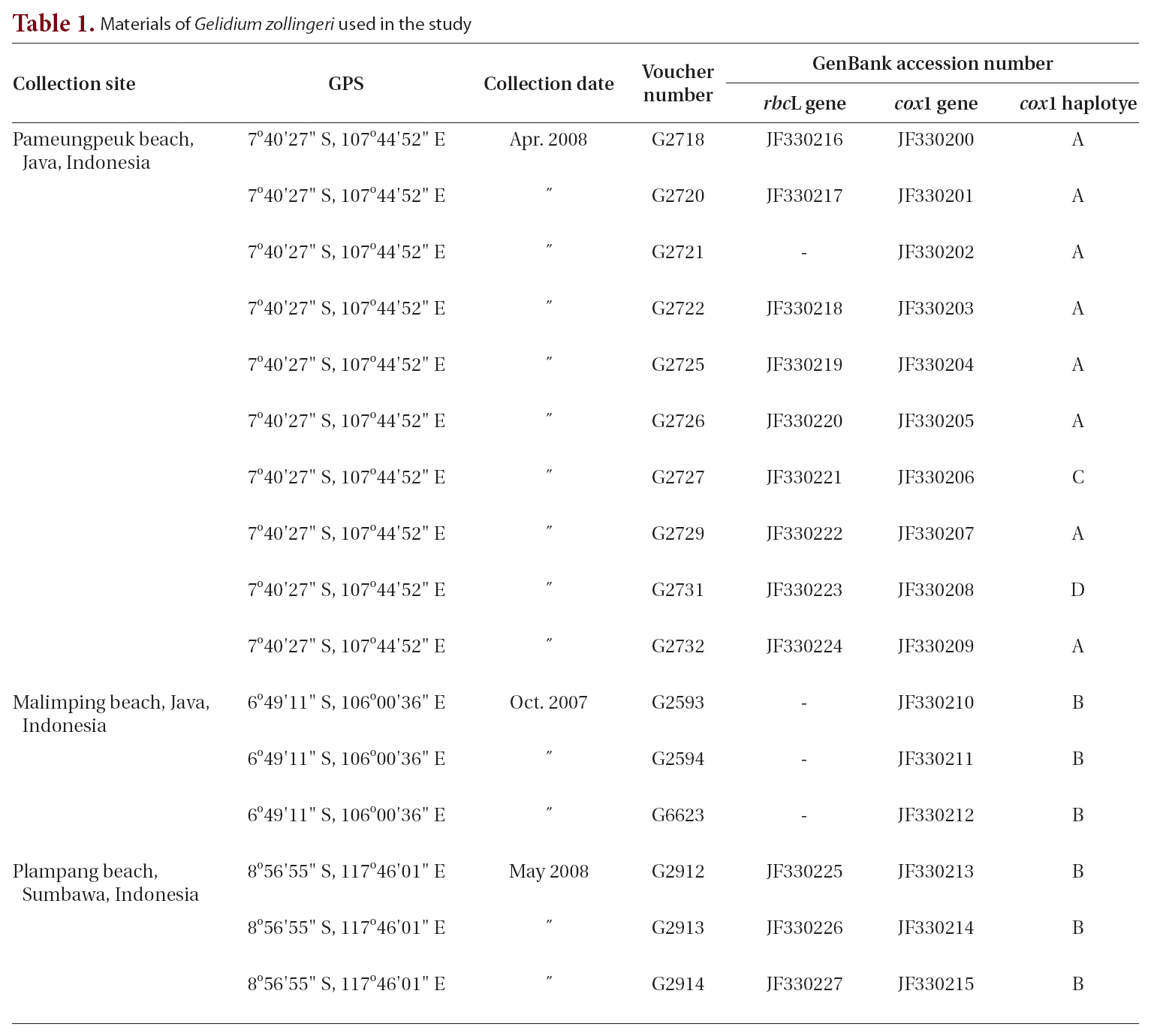
Sixty-eight sequences from 59 taxa of Porphyroglossum, Gelidium, and other genera within the family Gelidiaceae were aligned using a 1,266-nucleotide portion of rbcL. Variable sites occurred at 484 positions (38.2%), and 413 positions (32.6%) were parsimony-informative. P. zollingeri collections from Java Island in Indonesia formed a single monophyletic group with maximum support. P. zollingeri was sister to G. floridanum (51% for ML and 61% for MP) (Fig. 2). The genera Gelidium species and Porphyroglossum formed a single well supported clade (100% for ML and 95% for MP).
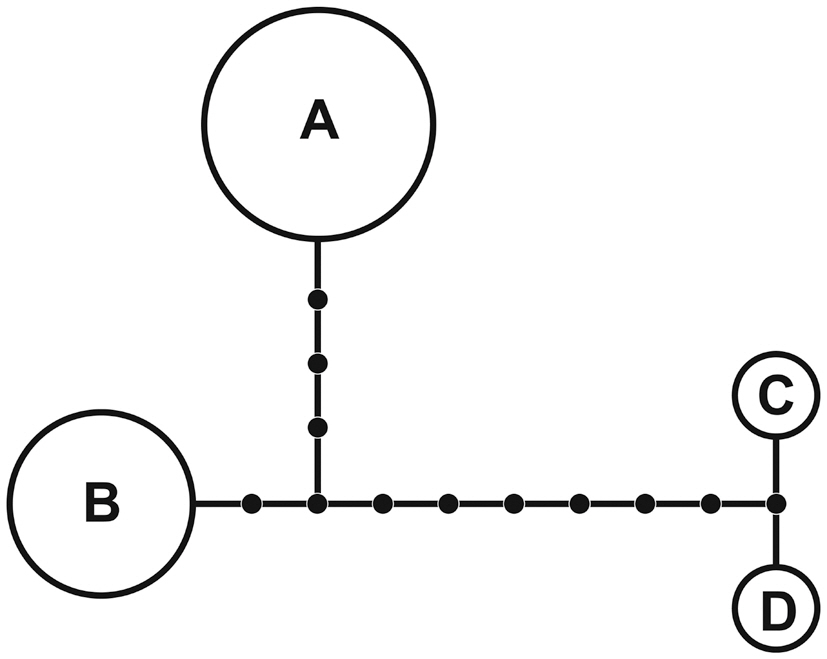
A 1,260-nucleotide portion of the cox1 gene was compared across 37 taxa of Porphyroglossum, Gelidium, Acanthopeltis, Gelidiella, and Pterocladiella. Variable sites occurred at 454 positions (38.4%), and 417 positions (31.1%) were parsimony-informative. Sixteen samples of P. zollingeri were used for haplotype analyses of cox1. The nucleotide and haplotype diversities were 0.004 and 0.642, respectively. The statistical parsimony network revealed four haplotypes (Fig. 3). Haplotype A was found in eight specimens and haplotype B occurred in six specimens.
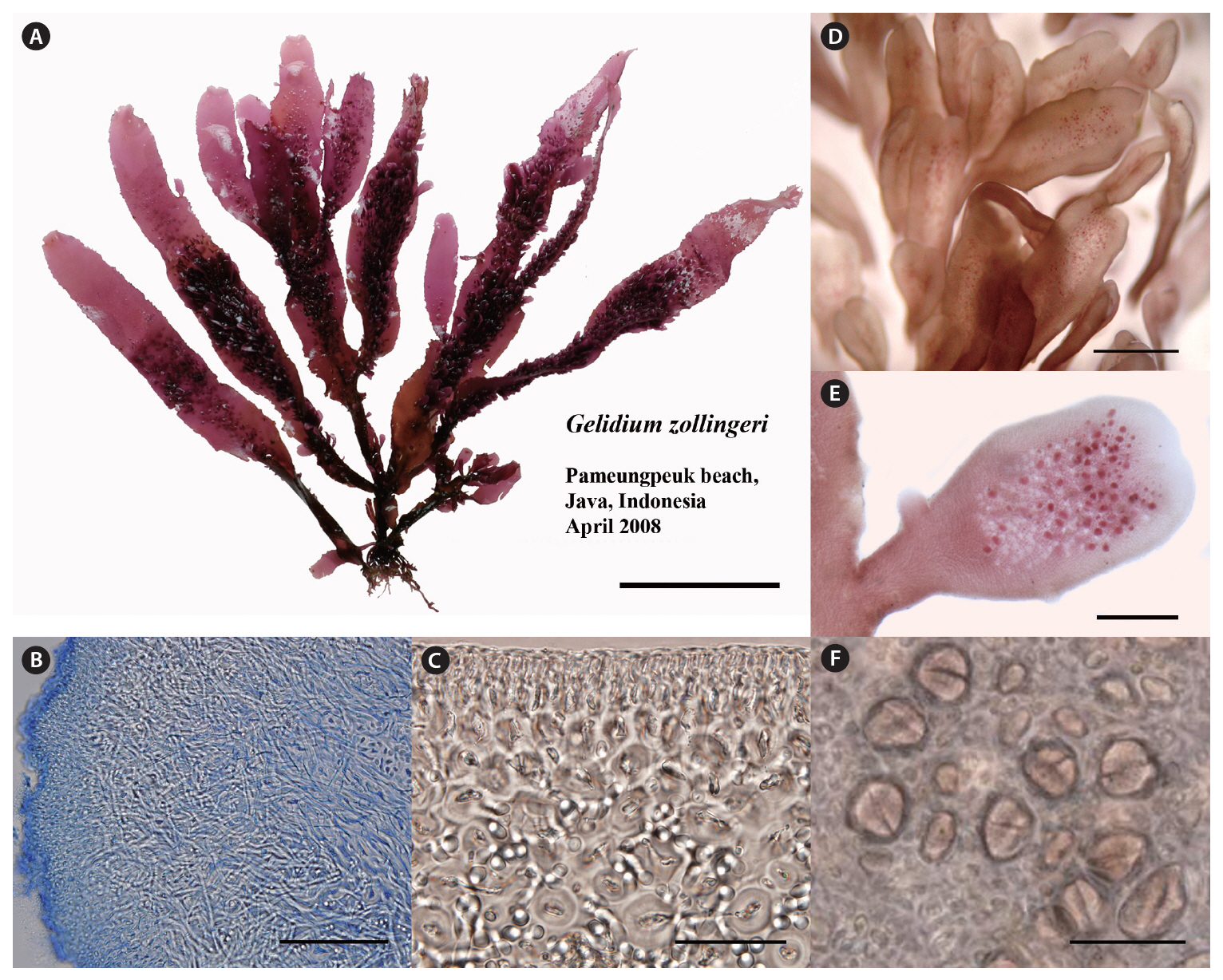
Thalli are cartilaginous, somewhat complanate when erect axes and branches are arranged in one plane, up to 15 cm high, arising from branched, recurved, stolon-like axes (Fig. 4A), and attached to the substratum by numerous peg-like haptera up to 0.5 mm in length. In a surface view, the cortical cells are rounded to ovate, mostly arranged in tetrads, 4-7 ㎛ in diameter. In cross section, the outer cortical cells are rounded, 4-8 ㎛ in diameter, gradually grading into larger cortical cells and then into medullary cells, 5-12 ㎛ in diameter. Rhizoidal filaments are abundant and interwoven in the longitudinal view (Fig. 4B). They are elongate and colorless, and concentrate around medullary cells (Fig. 4C). Erect axes are cylindrical at the base, progressively flattened above, ribbonlike in shape, up to 6 mm in width, ending in lanceolate, blunt, or truncate apices. The truncate ends often have ribbonlike proliferations similar in shape to the main axis but narrower. They arise in groups of two to four. The surfaces of main axes and proliferations are invested with longitudinally arranged rows of determinate, spatulate ramuli up to 1.5 mm in length (Fig. 4D). Tetrasporangial sori arise on small, determinate ramuli (Fig. 4E). Tetrasporangia are irregularly arranged in sori, rounded in surface view, and are 18-25 ㎛ in diameter (Fig. 4F). Mature tetrasporangia are cruciately divided.
This is the first report of gene sequences from Porphyroglossum, although many previous molecular analyses of other gelidioid algae have been performed (e.g., Freshwater and Rueness 1994, Freshwater et al. 1995, Shimada et al. 1999, Kim et al. in press). Through two-gene analyses and detailed morphological observations, we answered a question posed by Hatta and Prud’homme van Reine (1991) on the close relationships between Porphyroglossum and Gelidium. Phylogenetic analyses of plastid rbcL and mitochondrial cox1 sequences indicate that Porphy-
roglossum and Gelidium are not separate monophyletic groups. In all analyses of two individual genes and combined data, Porphyroglossum consistently nested within Gelidium, implying that P. zollingeri likely diverged from a common ancestral species of Gelidium.
Our descriptions of morphology and reproduction in P. zollingeri are in agreement with those of previous studies (Weber-van Bosse 1921, Fan 1961, Akatsuka 1983, Hatta and Prud’homme van Reine 1991). One of the distinguishing features of P. zollingeri is the presence of abundant proliferations on broad axes (Fig. 4A & D). However, surface proliferations also occur in Ptilophora (Tronchin et al. 2003) and they cannot be considered autapomorphic for Porphyroglossum. Notably, rhizoidal filaments are so abundant and interwoven in the medullary layer that it makes discerning medullary cells difficult, a rare phenomenon in most species of Gelidium.
We found no cystocarpic thalli in our collections of P. zollingeri. However, according to previous studies (Weber-van Bosse 1921, Fan 1961, Akatsuka 1986), cystocarps are bilocular with ostioles on both surfaces in flat or raised positions. The cystocarps are not different from those of Gelidium (Hatta and Prud’homme van Reine 1991). Male thalli have not been found in this species. Tetrasporangial characters of P. zollingeri in the present study are within the range of those in Gelidium.
On the basis of rbcL and cox1 sequences data and inconsistencies in key morphological characters distinguishing Porphyroglossum from Gelidium, we conclude that their maintenance as separate genera is untenable. Accordingly, we propose that Porphyroglossum Kutzing (1847) be merged with Gelidium Lamouroux (1813).
Gelidium zollingeri is related to G. floridanum with moderate support (51% for ML and 61% for MP) in the rbcL tree, and to G. serrulatum without support. Determining which characters are synapomorphic for these species is difficult. However, they are well resolved in the largest clade within Gelidium (100% for both ML and MP) comprising G. elegans, G. pacificum, G. linoides, G. tenuifolium, G. allanii, G. koshikianum, G. purpurascens, G. robustum, G. americanum, G. amansii, G. abbottiorum, G. pteridifolium, and G. proundum. This clade (with the exception of G. zollingeri) has been identified in previously published molecular studies (Freshwater et al. 1995, Millar and Freshwater 2005, Nelson et al. 2006, Kim et al. in press).
Network analysis of four cox1 haplotypes of G. zollingeri revealed distantly related clusters, with connections of one (between haplotypes C and D) to 11 (between haplotypes A and C or D) missing haplotypes (Fig. 3). We believe that the high number of missing haplotypes may be an artifact of sampling and that an increased sampling effort from areas with isolated haplotypes will significantly reduce the number of steps linking the clusters. However, four haplotypes and a high number of missing haplotypes in a very limited area of sampling in Indonesia suggest that high haplotype diversity is likely. Haplotype diversity of the cox1 gene is also high in Gelidiella fanii S.-M. Lin (Wiriyadamrikul et al. 2010).
In summary, of nine genera within the family Gelidiaceae, only three (Capreolia, Gelidium[including Acanth opeltis, Onikusa, Suhria, Porphyroglossum, Yatabella], and Ptilophora[including Beckerella]) remain as phylogenetically independent entities based on molecular marker(s). Phylogenetic studies using molecular markers have resulted in a broader concept of the genus Gelidium.The evolution of diverse morphologies, from cylindrical and compressed thalli (most species in Gelidium) via broad blades (G. japonicum, G. zollingeri) to sympodially growing axes with spirally arranged leaflike structures (species in the genus Acanthopeltis), may be the next focus of research on the genus Gelidium.
The formal synonym of Porphyroglossum follows: Gelidium zollingeri (Kutzing) K. M. Kim, G. S. Gerung and S. M. Boo comb. nov. Basionym: Porphyroglossum zollingeri Kutzing 1847: 775. Type: SE Java near Malang, Zollinger 2109 (L 941, 182-77).




![Maximum likelihood tree of 68 rbcL sequences calculated using the GTR + Γ + I evolution model (-lnL = 10893.760045; substitution rate matrix RAC = 1.184322 RAG = 8.436870 RAT = 1.973412 RCG = 1.752931 RCT = 14.046278 RGT = 1; base frequencies πA = 0.306815 πC = 0.163813 πG = 0. 216361 πT = 0.313011; shape parameter [α] = 0.199229). Maximum likelihood and maximum parsimony bootstrap values are shown for each clade. Figures in parenthesis refer to the number of specimens with identical sequences. AU Australia; BR Brazil; CL Chile; CR Costa Rica; ES Spain; FR France; IE Ireland; IT Italy; JP Japan; KR Korea; MA Morocco; MX Mexico; NA Namibia; NZ New Zealand; NO Norway; OM Oman; PH Philippines; TH Thailand; TW Taiwan; UK United Kingdom; US United States; VE Venezuela; ZA South Africa.](http://oak.go.kr/repository/journal/10663/JORHBK_2011_v26n1_33_f002.jpg)