Molecular genetic tools are widely used to learn more about the identical characterization of obscure microalgal strains. At the Korea Marine Microalgae Culture Center (KMMCC), the authors deduced the genetic relationship of 41 strains of the genus Tetraselmis by analysing a small subunit ribosomal DNA (18S rDNA) sequences. Forty-one strains were seperated into five groups, which showed over a 98-99% similarity to Tetraselmis striata or Tetraselmis sp. Tsbre. Also, 13 strains among them had an identical genotype to Tetraselmis striata while 5 strains had with Tetraselmis sp. Tsbre, respectively. The mean size of each strain generally showed the tendency of different variation according to the groups.
Species of
Since the first description of
Although, by using light and electron microscope, morphological classification is possible at the species level, taxonomic decisions within the
Genetic data based upon the amplification and sequencing of genes are also accumulated to use as a powerful tools for analysis of diverse microalgae. Thus, it is necessary to collect molecular data on
In this study, 18S rDNA sequence of 41 strains of
>
Microalgae culture condition
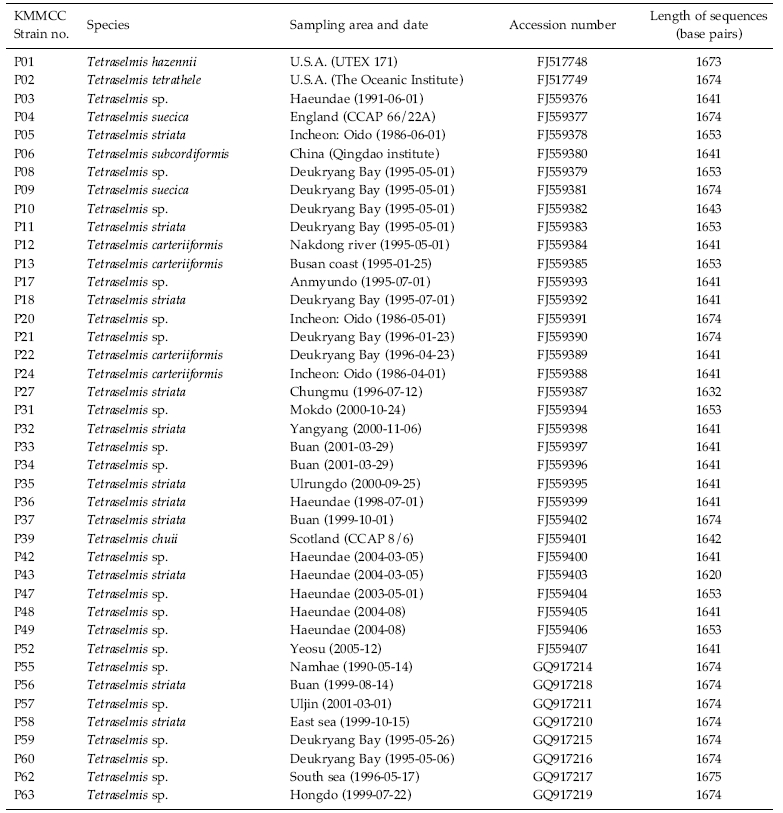
Forty-one strains of
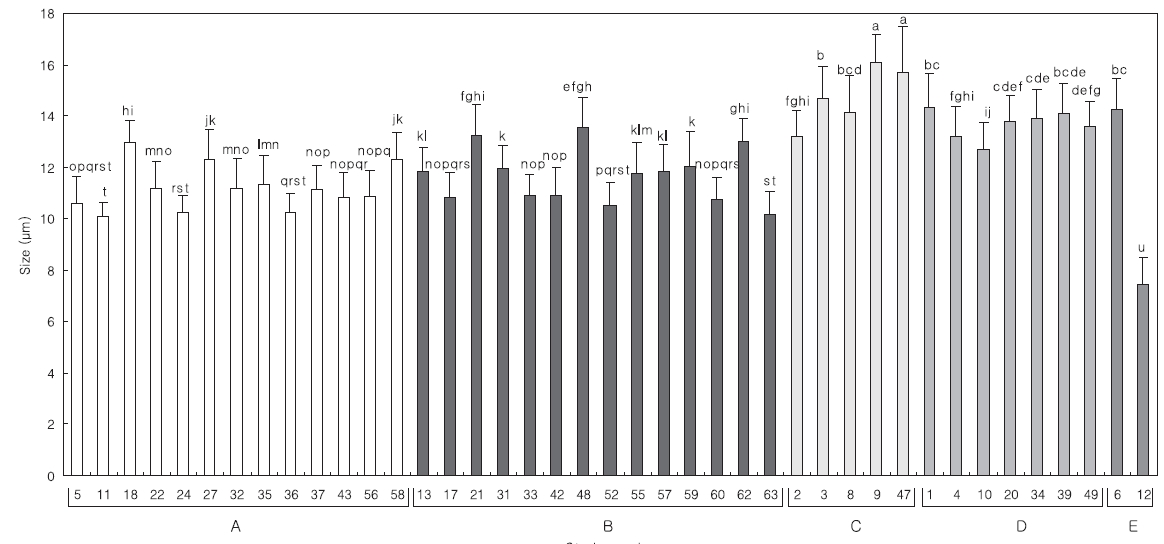
The Mean length of 30 specimens of the
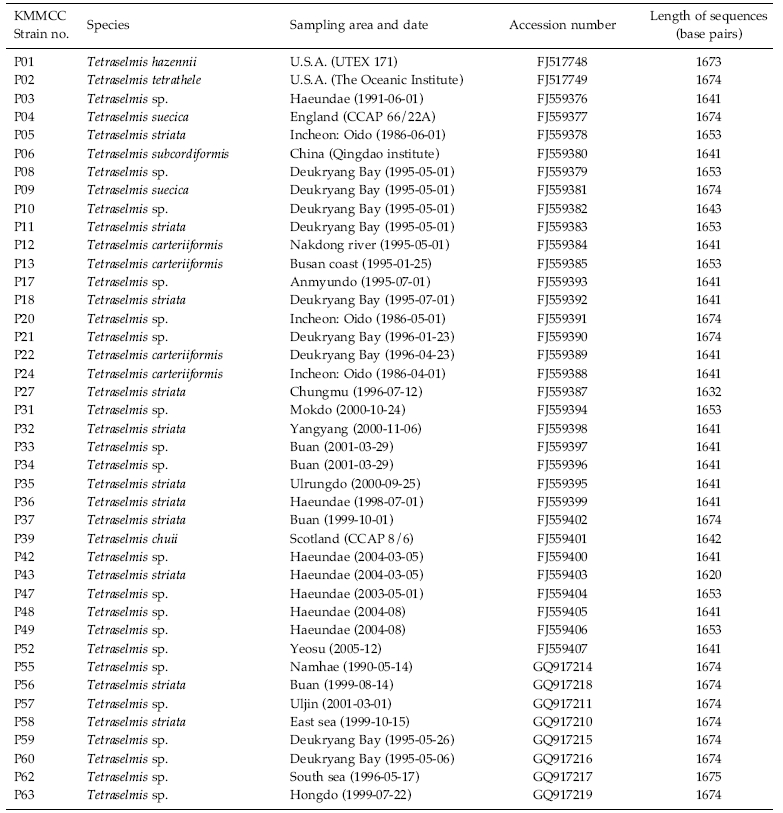
Culture history of forty-one strains of Tetraselmis and their GenBank accession numbers for the 18S rDNA sequences
>
Genomic DNA extraction and polymerase chain reactions
The LiCl method was used to extract the genomic DNA of the
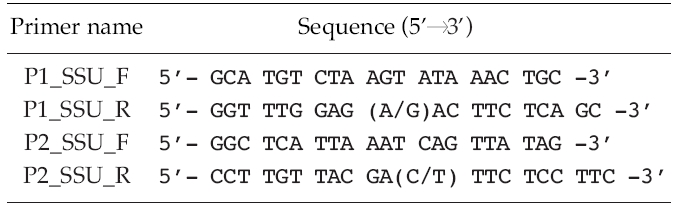
Polymerase chain reactions (PCR) (Mullis and Faloona 1987) performed using 10-100 ng of genomic DNA as a template and 0.5 μM of degenerated primers (Table 2) derived from a conserved region of 10 species of other known microalgae, listed in Table 3. Amplification conditions consisted of one cycle of denaturation at 95°C for 5 min, 30-35 cycles of denaturation at 95°C for 30 s, annealing at 50-55°C for 30 s and extension at 72°C for 1 min 45 s, followed by the final extension at 72°C for 7 min, and then stored at 4°C. About a 1.7 Kb size of PCR product was confirmed at 1% gel electrophoresis and extracted using the AccuPrep® Gel Purification Kit (Bioneer, Daejeon, Korea). Purified PCR products were ligated with the pGEM®-T Easy Vector Systems (Promega, San Luis Obisco, CA, USA) and then were transformed into the
[Table 2.] List of sequences of oligonucleotides used for the polymerase chain reactions
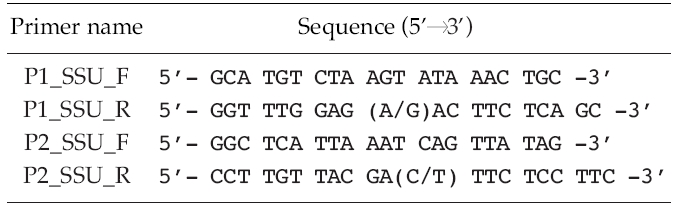
List of sequences of oligonucleotides used for the polymerase chain reactions
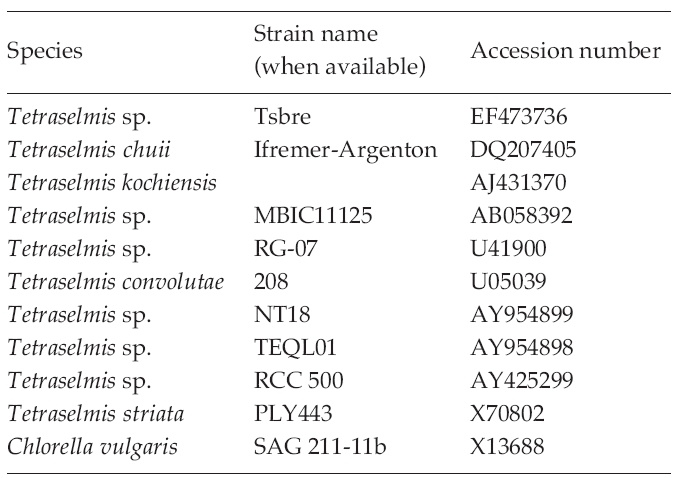
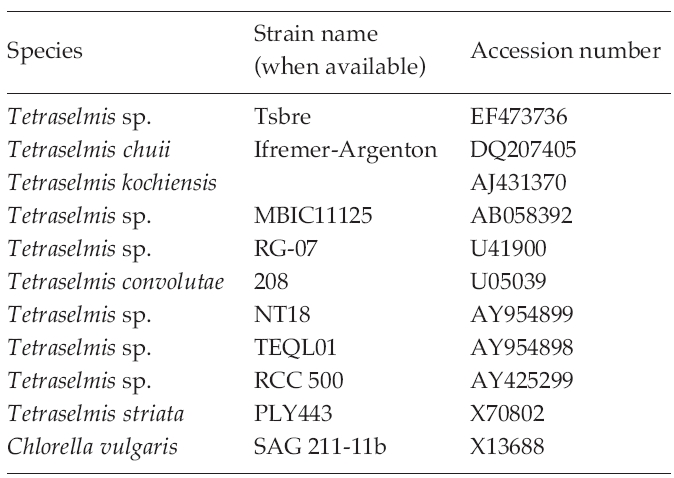
The strains used for the construction of a phylogenetic tree and their GenBank accession numbers of 18S rDNA sequences
>
Sequence alignments and phylogenetic analysis
The identities of the acquired partial 18S rDNA sequence from
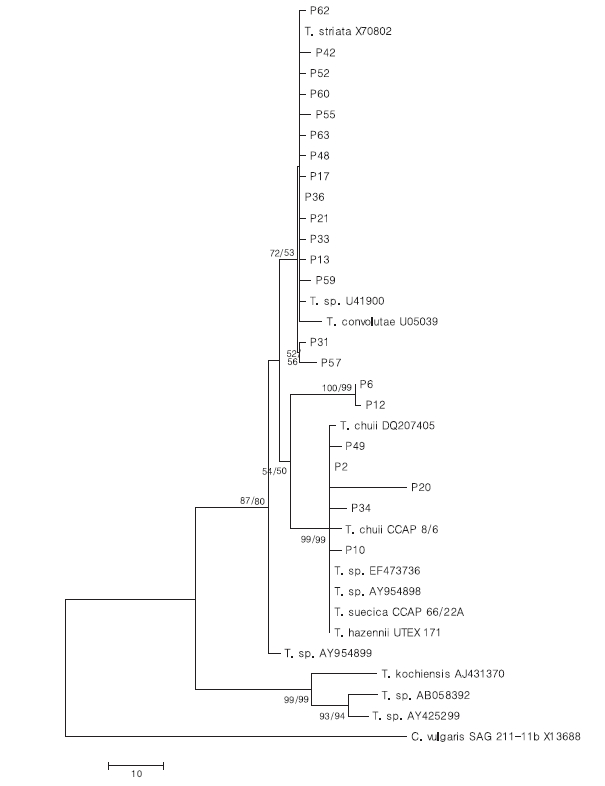
To confirm the phylogenetic relationships, a total of 1615 positions in the final dataset were subjected to neighbor-joining (NJ) and maximum-parsimony (MP) analysis using the MEGA v.4.0 (MEGA, Tempe, AZ, USA) (Tamura
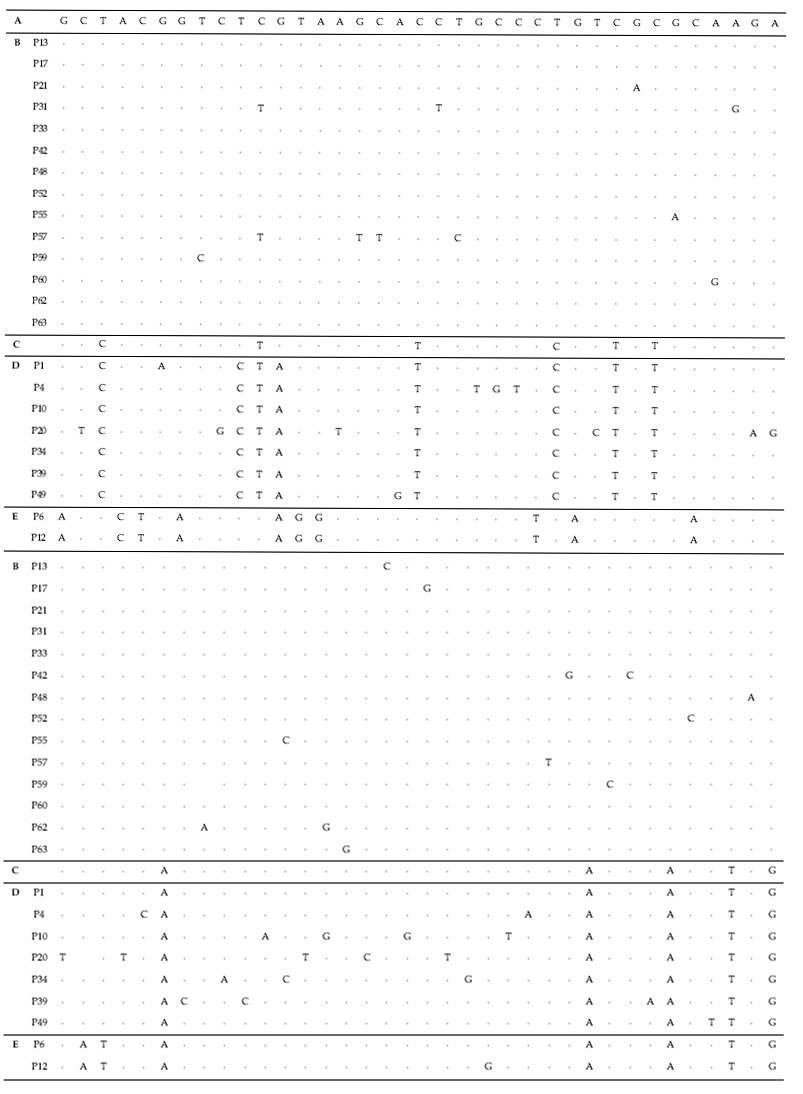
To test their generic similarity, the almost complete 18S rDNA sequences were determined for the 36 strains collected from Korean coastal water and 5 strains from foreign areas. Their accession numbers registered at GenBank are given in Table 1 and the lengths of sequence were varied on the sense and antisense primer used in gene amplification (Table 2). Forty-one strains showed high levels of identity (98-99%) to the correspoding regions of known 18S rDNA sequences such as
The sequences of the other 35 strains were seperated into 3 groups; B, D and E. Of these, fourteen strains belonged to group B, which was very close to group A. Only 1-2 sequence positions differentiated between group A and B, excepting KMMCC P-57. Group D contained seven strains which were close to group C but the level of similarity between groups C and D was less so than between groups A and B. Different sequence positions of six strains of group D were 1-4 base pairs compared with group C, whereas KMMCC P-20 was the furtherest from it with 14 sequence differences. The different sequence position of KMMCC P-6 and P-12 in group E showed only one base pair. But group E differed slightly from both of group A and group C. It had 17 different sequence positions in KMMCC P-6 or 18 differences in KMMCC P-12 compare with group A. Eighteen or 19 base pairs differed between group C and KMMCC P-6 or P-12, respectively.
Comparing the algal size with the 18S rDNA sequence analysis in each group, the relationship exhibited more clearly in the algal length than in the width (data not shown). The distribution of mean length is divided into; groups A and B, and groups C and D. The length of the strains from group B was similar to that of group A,
To analyze the relationship of 18S rDNA sequences among the strains of
One of the objectives of this research on their rDNA sequences was to identify strains of