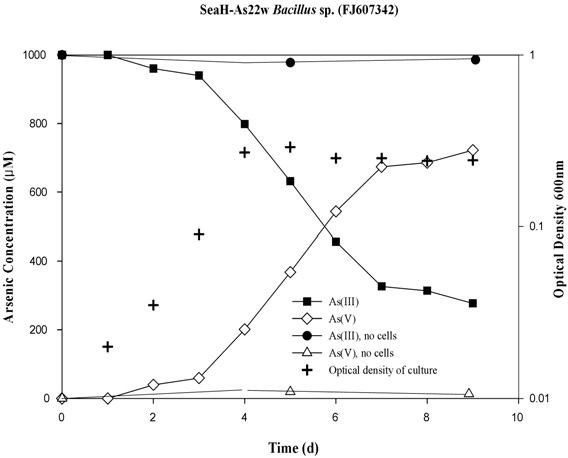
This study was conducted to evaluated seawater bacteria and their seasonal characteristics in the arsenic contaminated coastal seawater of Yeosu Bay, the Republic of Korea. Arsenite-oxidizing bacteria play an important role in the seawater of the arsenic contaminated bay, with a variety of arsenic resistance system (ars) genotypes being present during summer. Specifically, Bacillus sp. strain SeaH-As22w (FJ607342), isolated from the bay, were found to contain the arsB, arrA and aoxR type operons, which are involved in arsenic resistance. The isolated bacteria showed relatively high tolerance to sodium arsenite (III; NaAsO2) at concentrations as high as 50 mM. Additionally, batch seawater experiments showed that Bacillus sp. strain SeaH-As22w completely oxidized 1 mM of As (III) to As (V) within 10 days. Ecologically, the arsenic-oxidizing potential plays an important role in arsenic toxicity and mobility in As-contaminated coastal seawater of Yeosu Bay during all seasons because it facilitates the activity of Bacillus sp. groups.
Arsenic compounds are ubiquitous in nature and commonly found in environmental samples involved in many different biological-chemical interactions, including arsenite-oxidation[1- 3]. Arsenic compounds naturally occur in marine systems and are concentrated in a variety of ores[2, 4]. Arsenic in marine systems is predominantly in inorganic forms, such as arsenite (As (III)) and arsenate (As (V)), and organic forms, such as methylarsenate (MA), dimethylarsinate (DMA), trimethylarsine oxide (TMAO), teramethylarsonim ion (TMA), arsenocholine (AC) and arsenobetaine (AB). A study of arsenic in an aquatic system was conducted by Atkins and Wilson in 1927, and their results were similar to these reported for generally inconsiderable higher than those in seawater[4]. To date, seawater samples have provided the greatest number of marine arsenic compounds, although recent studies have revealed that terrestrial samples also contain many of these compounds. A significant percentage of the arsenic in surface water from marine environments is oxidized to arsenate, and is then methylated[2]. Previous studies have shown that the arsenic-resistant bacteria isolated from marine environments, are able to accumulate arsenic[6, 7]. Furthermore, arsenic cycling has been shown to respond to biological redox in seawater as well as in the demethylation assay.
It is well known that arsenic redox transformations by microorganisms are mediated by specific enzymes or respiratory chains[8]. The results of these studies suggest that arsenic species change in response to bacteria with the
In this study, arsenic-resistant bacterial strains were isolated from the eastern coastal seawater of Yeosu Bay during spring, summer, autumn and winter, and then identified based on their
2.1. Isolation and Growth of Arsenic Oxidizing Bacteria
Seawater samples were collected for isolating pure bacteria from the Yeosu Bay, Republic of Korea. To isolate arsenicresistant microorganisms from the samples collected during spring, summer, autumn and winter (Fig. 1), 1 mL of seawater was added to MSB medium[8, 9] containing sodium arsenite (NaAsO2) (Sigma, USA). After several transfers, isolated colonies were assessed for the amounts of As (III) present, with a single isolate selected for further study. The isolates (7th pass) were then cultures on media containing sodium arsenite at concentrations ranging from 0 to 26 mM (As (III): NaAsO2; 0, 5, 10, 15, 20 and 26 mM). The isolated arsenic resistant bacteria were tested for their ability to oxidize As (III). The arsenite-oxidizing microorganisms collected during spring, summer, autumn and winter were cultured at 22℃ in MSB (Stranier'Basal Medium, pH 7), with 1 mM D(+)-glucose as the carbon source. This medium was then autoclaved at 121℃ for 25 minutes before use. Prior to inoculation, the bacteria were harvested (14 kg, 20 min). After incubation, colonies were removed using a sterile syringe, with anoxic conditions maintained by continuous flushing of the culture tube with 22℃ O2-headspace, with H2 (10% in N2) as the electron donor in lieu of As (III). The isolated colonies were again assessed for their levels of sodium arsenite (NaAsO2), after which a single isolate was selected. Next, arsenic resistance experiments were carried out using the same arsenic concentrations used in the aerobic tests. For anoxic liquid cultures, MSB (Stranier'Basal Medium, pH 7) was first preincubated in the glove box of a chamber for at least 3 days. The arsenic resistant bacterium grew in MSB plus (1mM) D(+)-glucose.
The temperatures and pH values were measured using an Orion model 290A portable meter, employing an Orion model 9170 electrode. To determine the total concentrations of arsenic, 25 mL of distilled water were added to each sample. The samples were then shaken, after which the water phase was filtered through a membrane PTFE filter (Whatman 0.45-μm pore size, 13 mm), with 1 mL of HNO3 and 3 mL of HCl added to 0.25 g of the residue. The samples were again filtered through a membrane PTFE filter (Whatman 0.2-μm pore size, 13 mm), after which the arsenic concentrations were measured using a hydride-generation atomic absorption spectrophotometer (HG-AAS, Perkin Elmer 5100, Waltham, MA, USA), with a detection limit of 1 μmol/L[12]. All seawater of samples were collected from depth of 1 ~ 5 m. Analyses of heavy metals and arsenic were conducted according to standard procedures[13, 14].
2.2. PCR Amplification of the 16S rDNA and ars Genes
Bacterial genomic DNA was prepared from the bacterial cultures using standard methods[9, 15]. after which they were placed in a 1 mL microcentrifuge tube, with an individual colony of bacteria, to a final concentration of 108 CFU/mL. Each culture was then incubated overnight at 22℃, with intermittent shaking, and then placed in a 1.5 mL microcentrifuge tube, with 1 mL of TES (10 mL Tris-HCl, 50 mM EDTA, 10% sodium dodecyl sulfate) and 10 μL of proteinase K (50 mg l-1), which was subsequently reacted in a 55℃ tremulous cistern for 10-12 hours to digest the protein. An equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) was added to the reaction mixture. This solution was then manually mixed for 3 minutes and centrifuged at 14,240 g for 15 minutes, after which the supernatant was removed. This process was repeated 3 times, after which the supernatants were combined and mixed with 0.1 volumes of 3 M sodium acetate (pH 5.2) and 2.5 volumes of 100 % ethanol. The tubes were then shaken slowly, with the genomic DNA gently removed and then washed with 75 % ethanol, dried, and dissolved in TE (10 mM Tris pH 8.0; 1 mM EDTA pH 7.2). The purity of the DNA was then determined based on the ratio of the absorbance at 260 and 280 nm, with only DNA with a ratio of 1.7 to 2.0 used for subsequent Bacterial genomic DNA was isolated from the cultures using standard methods[9, 15], and then placed in a 1 mL microcentrifuge tube with a single appropriate colony. The 16S rRNA gene was then amplified using oligonucleotide primers designed based on the published sequence. The sequences of each of the primers used to amplify the arsenic resistance gene are shown in Table 1. The PCR amplification was conducted by subjecting the reaction mixture, in a final volume of 50 μL, which contained 0.5 μg genomic DNA and 10 pmol of each of the primers, to 35 cycles of denaturation for 5min at 94℃, annealing at 55℃ for 1minute, and extension at 72℃ for 2 minutes, followed by an additional extension for 7 minutes at 72℃. Ars primers were designed to obtain a Tm of approximately 55 to 60℃, with specificity obtained by aligning the sequences using the BLAST Search Tool (BLAST; National Center for Biotechnology Information [http:www.ncbi.nlm.nih.gov]). The PCR was conducted in a Mastercycler Gradient (Eppendorf, Germany), with the PCR products analyzed in 0.7 to 1.5 % agarose gels (Sigma, USA). The ars primers were designed for the

Oligonucleotide primers used for amplification of the arsenic-resistant system genes Update sources: Chang et al.[8,9,11]
2.3. 16S rDNA Sequencing and Phylogenetic Analysis
The nucleotide sequences were determined using the dideoxychain termination method, with a PRISM Ready Reaction Dye terminator/primer cycle sequencing kit (Perkin-Elmer Corp., Norwalk, CT, USA). The oligonucleotide primers included in this investigation were specific for the 16S rDNA gene. Sequencing was conducted in a reaction mixture with a final volume of 6 μL, which contained 15 to 50 ng of the PCR product and 1 pmol of each primer. The samples were analyzed using an automated DNA sequencer (Model 3100; ABI PRISM Genetic Analyzer System Profile, USA), with the 16S rDNA sequencing results compared with sequences from the NCBI database. The BLAST algorithm, integrated with the Vector NTI Suite v5.5.1 (InforMax, USA), was used to determine the sequence homologies. Database sequences with fewer than 1,500 nucleotides were excluded from the phylogenetic analysis. The full 16S rDNA gene sequences were compiled using the Vector NTI Suite v5.5.1 (InforMax, USA). Phylogenetic analysis was conducted using the neighbor-joining method[16].
2.4. Arsenic Oxidation and Reduction Assays
To test the ability of the strains to oxidize arsenite and/or reduce arsenate, the isolates were inoculated in 250-mL glass flasks containing 60 mL of seasalts (Sigma, USA) and 1 mM sodium arsenite. For the batch tests, MSB medium, supplemented with 1 mM sodium arsenite, was inoculated with each strain (107/CFU), with the concentration of As (III) in the culture then determined. All experiments were conducted in triplicate, using 60-mL Erlenmeyer flasks; the bacteria were incubated aerobically at 22℃ for 10 d, with shaking (170 rpm). For controls, sterile medium (i.e., un-inoculated), with 1mM sodium arsenite, was incubated under the same conditions. Periodically, 2 mL samples were taken, and the cell density measured and the arsenic speciation determined. These samples were centrifuged at 14,240 g for 10 min, decanted, and then stored at 4℃ prior to arsenic analysis[12]. The oxidation of As (III) to As (V) throughout the incubation period was monitored by determining the concentration of As (III) based on the concentration of As (V), using a silica-based strong anion cartridge (LC-SAX SPE, Supelco)[17]. The As (III) and As (V) concentrations were then measured using a hydride generation atomic absorption spectrophotometer (HG-AAS, Perkin Elmer 5100). All analytical measurements were performed in duplicate.
3.1. Isolation and Characterization of Arsenic Oxidation by Seawater Microbial Strains
Arsenic was not found in the coastal seawater from Yeosu bay in every season; spring (AsIII; 15.4 μg/L±0.1, AsV;4.9 μg/L±0.1), summer (AsIII; 9.9 μg/L±0.1, AsV;12.9 μg/L±0.1), autumn (not detected) and winter (not detected) (Fig. 1). The concentration of arsenic required to influence biological-chemical interactions is known to range from 0.5 to 3 μg/L, with a mean of about 1.7 μg/L. [1] Additionally, arsenic is a suspected carcinogen and As-related cancer of the skin may occur following environmental exposure, such as the harvest of fish products from water containing arsenic at 0.0175 μg/L (at the 10-6 cancer risk level[18]. Arsenic contamination has a complex seawater biogeochemistry, with important implications of its toxicity towards organisms, including humans[19]. The arsenic concentrations observed during spring and summer in this study were high.
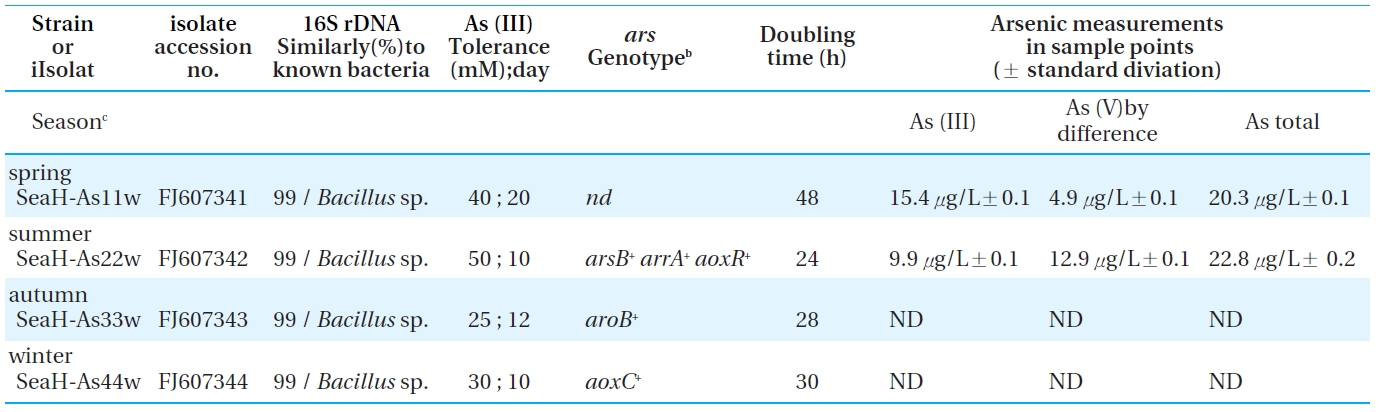
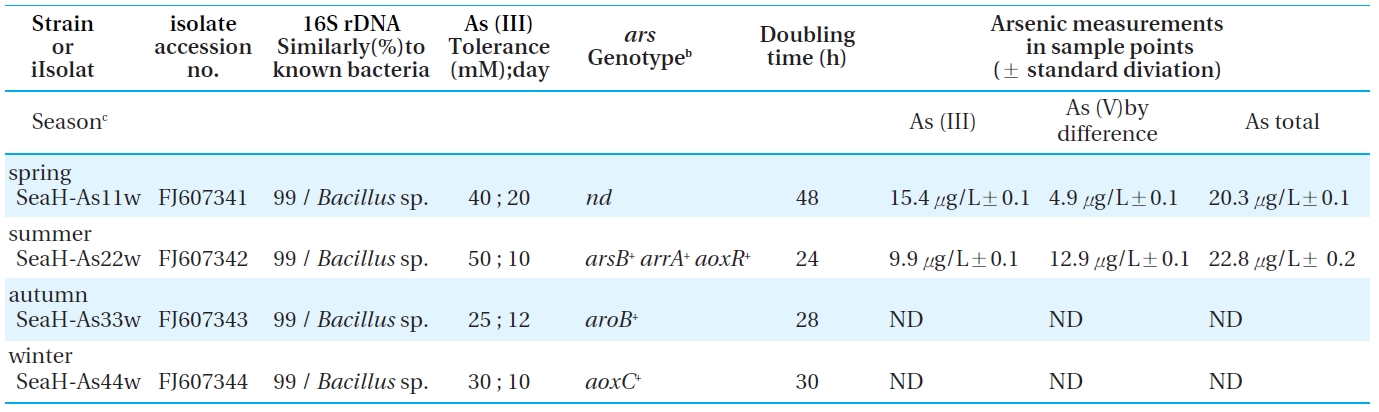
Bacterial strains, their identification, ars genotypes, and tolerance to arsenic under facultative anaerobic conditions. Each strain
Four indigenous arsenite oxidizing
Ecologically, arsenic-oxidizing potential plays an important role in As-detoxification/mobility in coastal seawater areas, as it facilitates marine-biogeochemical cycling activity of the
3.2. Sequencing of 16S rDNA and Phylogenetic Analysis
The phylogenetic relationships were analyzed based on the partial 16S rDNA sequences obtained from the 4 arsenic-resistant isolates, with Bacillales as an out-group, revealed all orders of the arsenic resistant Beta-proteobacteria. Four indigenous seawater bacteria strains were isolated; 2 As (III)-oxidizing bacteria and 2 arsenic resistant bacteria. According to our results, most isolates, except SeaH-As11w (FJ607341), SeaH-As22w (FJ607342), SeaHAs33w (FJ607343) and SeaH-As44w (FJ607344), were closely related to
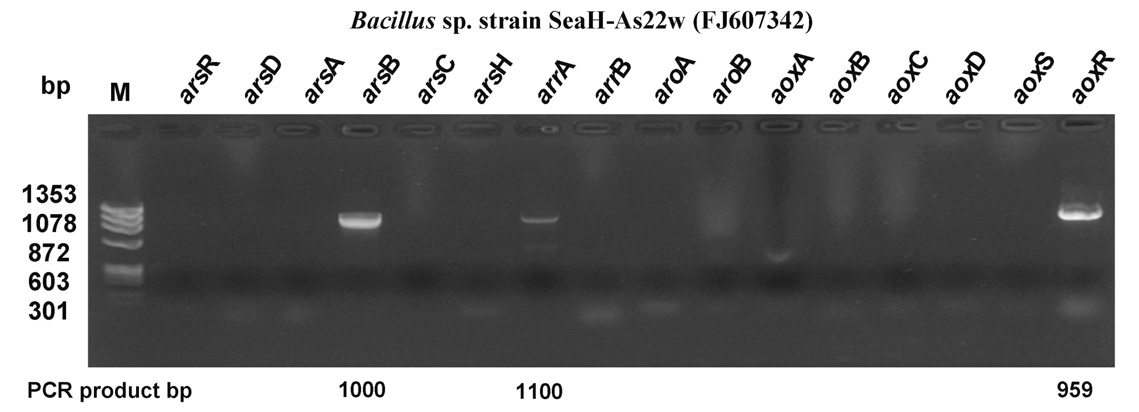
3.3. ars Genotype Profiling of Arsenite-Oxidizing Bacteria
The arsenic resistance system has been described based on an evaluation of the oxidation in the presence of bacteria containing the arsR, -D, -A, -AB, -B, -C, -H,
Herein is described the first investigation of As-oxidizing bacteria and
This work was supported by a grant (07SeaHeroA01-01) from the Plant Technology Advancement Program funded by the Ministry of Land, Transportation and Maritime Affairs of the Korean government.
![Oligonucleotide primers used for amplification of the arsenic-resistant system genes Update sources: Chang et al.[8,9,11]](http://oak.go.kr/repository/journal/10061/E1HGBK_2010_v15n1_15_t001.jpg)
![Phylogenetic tree based on the 16S rDNA sequence, showing the positions of the arsenite-oxidizing bacterial isolates SeaH-As22w (FJ607342) and Bacillales. The tree was constructed from a matrix of pair-wise genetic distances using the neighborjoining tree method. The phylogenetic data were obtained by aligning the different arsenic-resistant bacterial sequences in the Search Tool (BLAST; National Center for Biotechnology Information [http:www.ncbi.nlm.nih.gov]) using standard parameters. The scale bar represents 0.05 substitutions per 100 nucleotides within the16S rDNA sequence.](http://oak.go.kr/repository/journal/10061/E1HGBK_2010_v15n1_15_f003.jpg)