Periodontal disease is a chronic inflammatory infectious disease occurring in dental tissue supporting teeth1). It was reported that most of the adults are contracted by periodontal disease2) and prevalence of periodontal disease of domestic adults is showing a tendency of being increased continuously3). According to national health nutrients survey being performed by Centers for Disease Control in 2008, it was revealed that periodontal disease prevalence of our domestic adults was 73.9% and it was rapidly increased after the forties4). Periodontal disease induces diversified symptoms including gingival bleeding, swelling, formation of periodontal pocket, loss of attached gingiva and destruction of alveolar bone and it is a major cause of teeth loss5,6). Since the period from 1960s to 1970s when periodontal disease was proved to occur by oral bacteria infection7-9), dental plaque has been known as major cause of periodontal disease10).
Dental plaque means cohesive biofilm that is formed by various oral bacteria being attached to acquired pellicle being formed on dental surface11) and most of oral bacteria exist in a form of biofilm12,13). At an early stage of biofilm formation, Streptococcus gordonii is attached to host protein of dental surface and Streptococcus mutans, Streptococcus salivarius and Streptococcus mitis are found12). Afterwards, Veillonella and other anaerobic bacteria are also represented and Fusobacterium nucleatum is attached to early colony12,14). Bacterial distribution in oral biofilm varies depending on oral environmental elements such as pH, reduction potential and nutrition15). Therefore, the more oral environment is changed to anaerobic nature, the more number of anaerobic bacteria is increased14).
Anaerobic bacteria such as F. nucleatum, Porphyromonas gingivalis are frequently discovered in gingival lesion and they have relation with occurrence and progress of periodontal disease16,17). It was reported that amount, ratio and occurrence frequency of these anaerobic bacteria were increased in subgingiva of patients with chronic periodontal disease18) and in deep probing site of Korean patients with periodontal disease also, bacterial occurrence frequency was represented over 80%19). Among these, P. gingivalis that is anaerobic gram negative bacillus inhibits phagocytosis and anti-bacterial activity of neutrophilic leukocyte by producing gingipain that is a protease decomposing collagen of periodontal tissue and creating cytotoxic20-24). In addition, Treponema denticola has mobility as spiral shaped, obligatory anaerobic and gram negative bacterium and it creates infection by being attached to and penetrating into host tissue or substrate protein through protease and adhesion13,20,25,26). F. nucleatum plays a role of bridge connecting later stage colony such as P. gingivalis, T. denticola, Tannerella forsythia with early stage colony like Streptococci species (spp.)27-29). In other words, F. nucleatum is attached to early colony attached to dental surface and on top of it, later stage colony is attached and then biofilm is formed27,28).
Most of bacteria existing in biofilm is able to be attached and cohere with other bacteria and through this reaction, it forms dental plaque and helps other bacteria’s attachment30). Cohesion among bacteria takes place among colonies at an early stage of dental plaque and between early colony and later stage colony and it was also reported that it could take place in later stage colony as well31). Um et al.30) reported that this cohesive reaction situates bacteria closely and mutual interaction could be facilitated.
Interaction in bacterial colony is represented by various phenomena and it affects growth of different strains through nutrients production as well7,13,32). Lactic acid being produced by Streptococci and Actinomyces is used by Veillonella spp. as its energy source7,13). In addition, Vitamin K produced by Veillonella parvula is used at the time of Porphyromonas spp. growth and isobutyrate that is celluar membrane fatty acid produced by Fusobacterium spp. is used by Treponema microdentium32).
This interaction among bacteria is helpful for symbiosis but antagonism often takes place in some bacteria existing in dental plaque13,33). Streptococci inhibits proliferation of Aggregatibacter actinomycetemcomitans by producing hydrogen peroxide13) and A. actinomycetemcomitans inhibits growth of Streptococcus sanguinis in dental plaque by producing bactericin7). Like this, distribution of bacteria in biofilm also takes place by diversified interaction of bacteria34,35). Therefore, it could be explained that oral bacterial distribution is changed by inter-bacterial factors1).
A lot of researches on oral inter-bacterial interaction that creates periodontal disease have been performed but a study on an effect of interaction with dead bacteria on bacterial colony formation is deficient in reality.
Therefore, in this study, an effect of dead bacteria of S. gordonii, F. nucleatum and P. gingivalis on bacterial colony formation was intended to be explored. In other words, in order to confirm colony formation level, absorbance was measured and in order to confirm colony structure and form, it was observed with scanning electron microscope. In addition, in order to confirm an effect on pathogenicity of P. gingivalis that is major bacterium causing periodontal disease, expression analysis for rgpA gene was performed by using real time reverse transcriptase polymerase chain reaction (RT-PCR). Through this, by clarifying an effect of inter-bacterial interaction, a basic data required for providing a method of being able to prevent periodontal disease effectively is intended to be provided.
Strains used for this study are S. gordonii KCTC 5640, F. nucleatum KCTC 2640 and P. gingivalis KCTC 5352 and it was used by purchasing it from Korean Collection for Type Cultures (KCTC) in Korea Research Institute Bioscience and Biotechnology (KRIBB).
20 μl of S. gordonii, F. nucleatum and P. gingivalis were dispensed in 2 ml of sterilized trypticase soy broth with hemin and menadione (TSB; MBcell, Los Angeles, CA, USA) and cultured in 37°C CO2 incubator (Mco-175; Sanyo Electric Co., Ltd., Tokyo, Japan) under anaerobic gas pack condition for 7 days through anaerobic jar and all the strains used for this study were sub-cultured for more than 2 times.
In order to dispense based on same quantity, the number of bacteria of subcultured strains was measured by using UV/Vis Spectrophotometer (DU 730 Life Science UV/Vis Spectrophotometer; Beckman Coulter, Indianapolis, IN, USA). Afterwards, each of the bacteria (1×107 bacteria/ml) was cultured on a single basis for 7 days by making it suspended in TSB. In addition, in order to explore an interaction with dead bacteria, co-culture for such strains was performed based on same amount (1×107 bacteria/ml) as that of bacteria being cultured on a single basis after heat treating for 30 minutes at 100°C.
In order to explore colony formation level at the time of single culture of each bacterium and co-culture with dead bacteria, absorbance was measured. After mixing each colony evenly by vortex, it was dispensed to 96 well microtitier plate by each 200 μl and absorbance was measured at 595 nm by using microplate reader (GENios Spectra Fluor Plus; Tecan Group, Männedorf, Switzerland).
In order to confirm colony structure and form being formed by single culture of each bacterium and co-culture with dead bacteria, it was observed by scanning electron microscope. Each bacterium was pre-fixed in 2.5% glutaraldehyde (in 0.1 M phosphate buffer solution [PBS], pH 7.4) for 4 hours after washing it with PBS (0.1 M PBS, pH 7.4) for 2 times (10 minutes/time). Afterwards, it was post-fixed in 1% osmium tetroxide (in 0.1 M PBS, pH 7.4) for 1 hour after washing it with same PBS for 2 times (10 minutes/time). Again, after washing it with same PBS for 1 time (10 minutes/1 time), it was dehydrated in the order of ethanol concentration increase. After dehydration is completed, it was dried by using critical point dryer (HCP-2; Hitachi, Ibaraki, Japan) and after performing platinum coating in Ion sputter (E-1030; Hitachi) and attaching specimen to stub, it was observed by scanning electron microscope (S-4700; Hitachi) under 10 kV.
In order to explore an effect of dead bacteria of other strains on pathogenecity of P. gingivalis that is major bacterium causing periodontal disease, real-time RT-PCR was performed for analysis of rgpA expression that is a gene relevant to HRgpA. First, after mixing bacterial culture solution evenly with vortex, TSB (bacterium culture solution) of 1 ml was dispensed in 1.5 ml Eppendorf tube and it was centrifuged based on 2,000g for 1 minute by using centrifuger (Centrifuge5424R; Eppendorf, Hamburg, Germany). After removing supernatant, bacterial culture solution of 1 ml was dispensed again and it was centrifuged based on 2,000g for 1 minute. After removing supernatant to maximum, adding Pre Lysis Buffer of 250 μl (iNtRON, Seongnam, Korea) and making it resuspended again with pipette, it was incubated for 3 minutes at 95°C. And then, it was incubated at room temperature for 5 minutes after adding easy-RED BYF solution (iNtRON) of 750 μl and mixing it with vortex for 15 minutes. Afterwards, it was incubated at room temperature for 5 minutes after adding chloroform of 200 μl and mixing with vortex for 15 seconds again. After centrifuging it for 15 minutes at 4°C based on 21,130g, supernatant of 400 μl was dispensed to new 1.5 ml Eppendorf tube. After adding same amount of isopropanol and mixing it well by inverting the tube for 7 times, it was incubated for 10 minutes at room temperature. Afterwards, supernatant was removed after centrifugation for 10 minutes at 4°C based on 21,130g and after adding 70% ethanol of 1 ml and mixing it well by inverting the tube for 7 times, it was centrifuged for 5 minutes at 4°C based on 21,130g. After removing supernatant to maximum, drying remaining RNA pellet and dissolving such pellet in RNase-free water of 20 μl, its concentration was measured by using NanoDrop (Thermo Scientific, Wilmington, DE, USA).
Afterwards, after matching final capacity as 20 μl by mixing total RNA of 1 μl, I-Green 2× qPT-PCR Mix (iNtRON) of 10 μl, qRT-PCR Enzyme Mix (iNtRON) of 0.4 μl, forward and reverse primer of 1 μl and DNase/RNase free water, it was reacted at 42°C for 15 minutes, 95°C for 10 minutes, 95°C for 15 seconds and 60°C for 60 seconds respectively and this was measured for 40 times repeatedly. Sequence of primer being used in real-time RT-PCR is as shown on Table 1 and reaction result was represented as cycle threshold (Ct) by measuring it in StepOnePlus real-time RT-PCR system (Applied Biosystems, Foster City, CA, USA) in real time. The experiments in this study were performed three times in triplicate, it was represented by representative data.
For statistical analysis of this study, PASW Statistics ver. 18.0 (IBM Co., Armonk, NY, USA) was used and its result was verified by one-way ANOVA. In addition, through Dunnett T3, post-analysis was performed and when p-value was below 0.05, it was deemed to be significant.
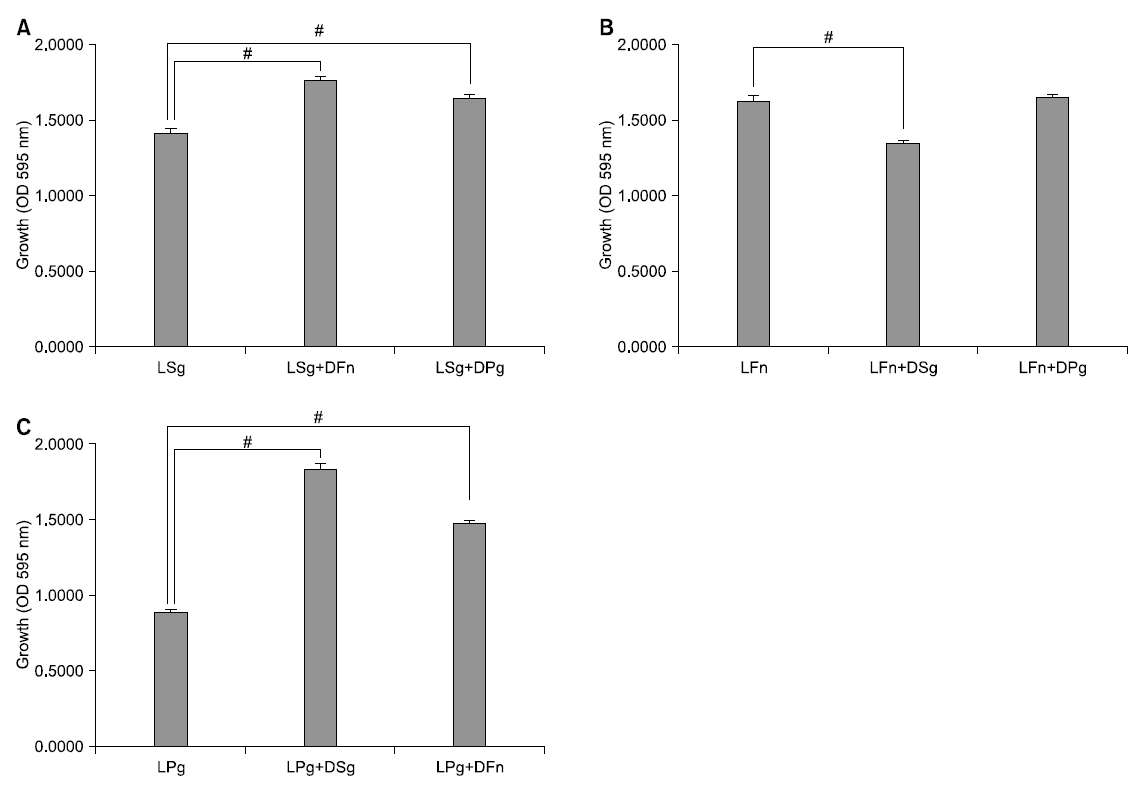
At the time of single culture of S. gordonii and co-culture with dead bacteria of other strains, growth was compared. At the time of single culture of S. gordonii, its absorbance was 1.420 and as a result of co-culture with dead F. nucleatum being killed with heat treatment for 30 minutes at 100°C, it was 1.773. In addition, in case of co-culture with dead P. gingivalis, it was 1.655. At the time of co-culture with dead bacteria, colony formation of S. gordonii was significantly increased statistically compared with single culture (p<0.05).
At the time of single culture of F. nucleatum and co-culture with dead bacteria of other strains, colony formation was compared. At the time of single culture of F. nucleatum, its absorbance was 1.637 and as a result of co-culture with dead S. gordonii being killed with heat treatment for 30 minutes at 100°C, it was 1.360. In addition, in case of co-culture with dead P. gingivalis, it was 1.660. At the time of co-culture with dead S. gordonii, growth of F. nucleatum was significantly decreased statistically compared with single culture (p<0.05). However, at the time of co-culture with dead P. gingivalis, it was not significant statistically compared with single culture but its colony formation was represented to be increased.
At the time of single culture of P. gingivalis and co-culture with dead bacteria of other strains, growth was compared. At the time of single culture of P. gingivalis, its absorbance was 0.890 and as a result of co-culture with dead S. gordonii being killed with heat treatment for 30 minutes at 100°C, it was 1.835. In addition, in case of co-culture with dead F. nucleatum, it was 1.483. When co-culture of P. gingivalis with dead bacteria, its growth was significantly increased than that of single culture statistically (p<0.05; Fig. 1).
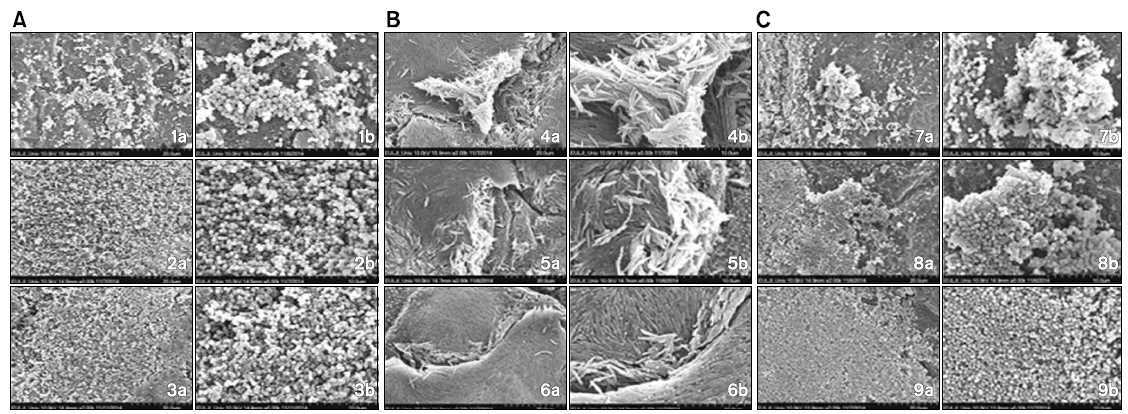
Structure and form difference between colony at the time of single culture and colony formation after co-culture with dead bacteria of other strains was confirmed by scanning electron microscope. At the time of co-culture with dead bacterium of F. nucleatum, colony formation of S. gordonii was more increased compared with single culture of S. gordonii. In addition, at the time of co-culture with dead bacterium of P. gingivalis also, colony of S. gordonii was observed to be more increased than that of single culture.
On the other hand, when co-culture of F. nucleatum with dead S. gordonii, colony formation of F. nucleatum was observed to be decreased than single culture but at the time of co-culture with dead P. gingivalis, its colony was more increased than that of single culture. In addition, at the time of co-culture of P. gingivalis with dead bacteria of other strains, it was confirmed that colony formation was more increased in both groups than that of single culture of P. gingivalis (Fig. 2).
[Fig. 2.] Scanning electron microscope images of colony structure and form being formed in single culture and co-culture. (A) Scanning electron microscope images of colony of Streptococcus gordonii with dead Fusobacterium nucleatum and Porphyromonas gingivalis. (B) Scanning electron microscope images of colony of F. nucleatum with dead S. gordonii and P. gingivalis. (C) Scanning electron microscope images of colony of P. gingivalis with dead S. gordonii and F. nucleatum. 1) Live S. gordonii, 2) live S. gordonii+dead F. nucleatum, 3) live S. gordonii+dead P. gingivalis, 4) live F. nucleatum, 5) live F. nucleatum+dead S. gordonii, 6) live F. nucleatum+ dead P. gingivalis, 7) live P. gingivalis, 8) live P. gingivalis+dead S. gordonii, 9) live P. gingivalis+dead F. nucleatum. a): ×2,000 times, b): ×5,000 times.
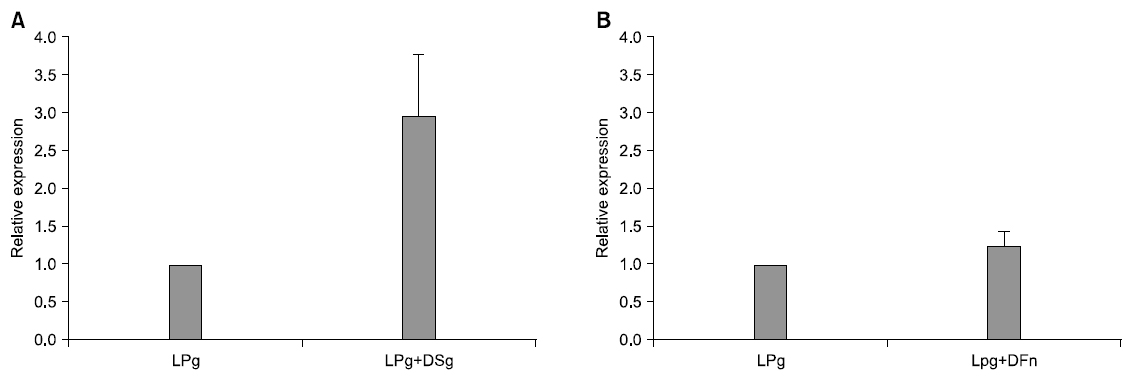
In order to analyze change of rgpA expression, that is gene relevant to HRgpA which is protease, of P. gingivalis being represented at the time of co-culture with dead bacteria, real-time RT-PCR was performed. Gene expression was quantified by comparing it with expression of P. gingivalis 16S. When co-culture with dead S. gordonii, its change was increased by 2.96 times than that of single culture. In addition, rgpA expression at the time of co-culture with dead F. nucleatum was increased by 1.23 times than that of single culture. RgpA expression that is gene of P. gingivalis was not significant statistically but when co-culture with dead S. gordonii and F. nucleatum, it was more increased than that of single culture (Fig. 3).
Various kinds of oral bacteria comprise biofilm12,36). S. gordonii is an early colony and it provides a place where bacteria causing periodontal disease could be attached to biofilm37). F. nucleatum helps bacteria causing periodontal disease attach to teeth or oral epithelial cells at the middle point of physical interaction of gram positive bacteria and gram negative bacteria. In addition, by decreasing immunity of host, it helps toxic mechanism relevant to other bacteria and plays a very important role in onset and progress of periodontal disease38). P. gingivalis, that is gram negative anaerobic bacillus, causes destruction of periodontal tissue by producing protease and delay of injury healing by impeding interaction between host cell and extracellular matrix38,39). It was known that periodontal disease is induced by complex interaction of these bacteria12,36). Therefore, in this study, an effect of interaction with dead bacteria on growth of S. gordonii, F. nucleatum and P. gingivalis that are bacteria relevant to periodontal disease was intended to be explored.
In this study, as a result of comparing growth at the time of single culture of S. gordonii and growth at the time of co-culture of S. gordonii with dead F. nucleatum, growth of S. gordonii at the time of co-culture with dead F. nucleatum was more significantly increased statistically than its single culture. When observing existing studies on interaction of live S. gordonii and F. nucleatum, in a study of Lee1) where each bacterium was separated by using film with 0.4 μm holes and cultured, they reported that at the time of co-culture with F. nucleatum, S. gordonii was significantly grown statistically. Jang40) also reported that at the time of co-culture with A1-2 (autoinducer-2) of F. nucleatum, biofilm of S. gordonii was more formed. In addition, in a study on proteomies change of S. gordonii at the time of development of oral biofilm, Hendrickson et al.41) also reported that when S. gordonii was present together with F. nucleatum rather than existing alone, energy metabolism was increased. On the other hand, as a result of this study comparing growth at the time of co-culture of S. gordonii and dead P. gingivalis, its colony formation was significantly increased statistically than single culture of S. gordonii. Lee1) reported that at the time of separated co-culture by using film with holes, growth of S. gordonii was facilitated by P. gingivalis. In addition, Hendrickson et al.41) also reported that at the time of development of oral biofilm, energy metabolism was increased when S. gordonii was present together with F. nucleatum rather than existing alone. In view of these results, it is considered that at the time of growth of S. gordonii, not only live bacteria but also dead bacteria of F. nucleatum and P. gingivalis provides synergy effect.
In this study, at the time of co-culture of F. nucleatum with dead bacterium of S. gordonii, its colony formation was more significantly decreased statistically than single culture. When observing existing studies on interaction of F. nucleatum and live S. gordonii, Lee1) reported that at the time of separated co-culture, S. gordonii impeded growth of F. nucleatum. In addition, based on this, they reported further that by inhibiting colony formation of F. nucleatum by using oral bacterial flora, oral environment could be maintained cleanly. On the other hand, as a result of this study comparing colony formation at the time of co-culture of F. nucleatum with dead P. gingivalis, at the time of co-culture of F. nucleatum with dead P. gingivalis, its result was not significant statistically than single culture of F. nucleatum but growth was increased. When observing existing studies on interaction of P. gingivalis for F. nucleatum, Lee1) reported that as a result of separated co-culture with P. gingivalis, the number of F. nucleatum was increased and was growing more rapidly than single culture as growth facilitating factor was secreted. Saito et al.42) also reported that when making AI-2 of P. gingivalis producing luxS gene inactivated, strengthening of biofilm formation of F. nucleatum was inhibited. In addition, they reported that biofilm formation of F. nucleatum was considerably facilitated and strengthened by protein molecule of P. gingivalis. In view of these results, it is considered that colony formation of F. nucleatum was inhibited by S. gordonii but P. gingivalis provides colony formation of F. nucleatum with synergy effect physically and chemically.
On the other hand, in this study, when comparing colony formation of single culture of P. gingivalis with colony formation of co-culture with dead S. gordonii, it was more increased in co-culture with dead bacterium than single culture. P. gingivalis is attached to S. gordonii mediated by pair of hapten-receptor. In addition, FilmA of P. gingivalis is bonded with glyceraldehyde-3-phosphate dehydrogenase on the surface of S. gordonii and short flimbriae is bonded with SspA and SspB (Antigen I/II) hapten of S. gordonii13). Kuboniwa et al.43) reported that when P. gingivalis is present with S. gordonii together in oral biofilm, it forms heterogeneous biofilm and this structure provides P. gingivalis with physiological support. Huang et al.37) also reported that under the environment of nicotine extract presence, heterogeneous biofilm of P. gingivalis and S. gordonii is frequently formed. In addition, in this study, when cultured P. gingivalis together with dead F. nucleatum, its colony formation was significantly increased than single culture. Lee1) reported that growth of P. gingivalis was increased by F. nucleatum. In addition, due to this, if one species is present in oral cavity, it would cause progress of periodontal disease and chronic periodontitis by facilitating growth of bacterium of the other species. Jang40) reported that AI-2 of F. nucleatum increased formation not only of bacterial membrane of P. gingivalis but also of heterogeneous bacterial membrane with other bacteria and cohesive reaction and by being inhibited by quorum sensing inhibitor, AI-2 plays an important role in interaction. In addition, Kuboniwa et al.43) also reported that P. gingivalis receives physiological support through heterogeneous biofilm formation with F. nucleatum. In view of these reports, it is considered that colony formation of P. gingivalis receives synergy effect from S. gordonii and F. nucleatum physically and chemically.
In this study, in order to confirm structure and form difference of colony being formed at the time of cultured each bacterium on a single basis and that being formed after co-cultured with dead bacteria of other strains, it was observed by scanning electron microscope. Comparing with single culture of S. gordonii, S. gordonii colony at the time of co-culture with dead bacterium of F. nucleatum was more increased. In addition, at the time of co-culture with dead P. gingivalis also, it was observed that S. gordonii colony was more increased than single culture. On the other hand, at the time of co-culture of F. nucleatum with dead bacterium of S. gordonii, F. nucleatum colony was decreased but at the time of co-culture with dead bacterium of P. gingivalis, F. nucleatum colony was more increased than single culture. In addition, at the time of co-culture of P. gingivalis with dead bacteria of other strains, its colony formation was confirmed to be more increased in both two groups than single culture. And its aspect was similar to that of optical density being tested in this study. Therefore, it is considered that not only metabolism product being synthesized when bacteria is proliferated and grown but also bacterial cell itself affects colony formation of other strains.
In order to observe what kind of effect was exerted on pathogenicity of P. gingivalis by other strains, gene expression change of P. gingivalis was confirmed through real-time RT-PCR. P. gingivalis induces cytokine regulation failure and functional damage of cells relevant to living organism defense system by producing gingipain which is trypsin-liked protease such as HRgpA, RgpB, Kgp. Among these, HRgpA is produced by rgpA gene and it increases blood penetration and destroys coagulated protein by using activity of kallikrein/kinin path relevant to gingivitis and blood coagulation system24). Therefore, in this study, as a result of confirming expression change of rgpA that is gene relevant to protease HRgpA of P. gingivalis, at the time of co-culture of P. gingivalis with dead S. gordonii, its expression frequency was not significant statistically but it was more increased than single culture of P. gingivalis. It is considered that the more biofilm is old, pathogenicity of P. gingivalis would be increased since this result would mean that as early attached S. gordonii becomes dead bacterium over time, it increases gingipain formation of P. gingivalis. In addition, at the time of co-culture with dead bacterium of F. nucleatum also, gene expression of rgpA was not significant statistically but it was more increased than single culture. Lee44) reported that at the time of co-culture of P. gingivalis with F. nucleatum, it induces more inflammable cytokine expression and production in monocytic strain. In addition, they reported that virulence of P. gingivalis is increased by communication with F. nucleatum and in lipopolysaccaride production also, it represents strong toxicity as lpxA and lpxD expression are increased. Jang40) also reported that at the time of co-culture of P. gingivalis with AI-2 of F. nucleatum, rgpA gene expression was more increased than single culture. Therefore, it is considered that F. nucleatum and S. gordonii would affect pathogenicity of P. gingivalis.
When summarizing the result of this study, colony formation of S. gordonii was increased by dead bacteria of F. nucleatum and P. gingivalis. In case of F. nucleatum, its colony formation was decreased by dead bacterium of S. gordonii but it had a tendency to increase its colony formation by co-culture with dead bacterium of P. gingivalis than that of single culture. In case of P. gingivalis, colony formation was increased by dead bacteria of F. nucleatum and S. gordonii. Therefore, it is considered that strains being used for this test would exchange its effect not only through interaction factors but also through bacterial cell itself at the time of colony formation. In the future, a study reflecting oral environment would be required and further study would be required as to by which substance and mechanism it is affected.
What kind of influence interaction of S. gordonii, F. nucleatum and P. gingivalis that are bacteria relevant to periodontal disease exerts on colony formation was intended to be explored. In other words, in order to confirm colony formation level, absorbance was measured and in order to confirm colony structure and form, it was observed by scanning electron microscope. In addition, in order to confirm an effect on pathogenicity of P. gingivalis, it was performed by real-time RT-PCR. Afterwards, the significance was verified. As a result of this study, colony formation of S. gordonii was increased by dead bacteria of F. nucleatum and P. gingivalis. In case of F. nucleatum, its colony formation was decreased by dead bacterium of S. gordonii but it was represented to be increased by dead bacterium of P. gingivalis. In case of P. gingivalis, colony formation was increased by both dead bacteria of F. nucleatum and S. gordonii. Therefore, it is considered that strains being used in this test would exchange its effect not only through interaction factors but also through bacterial cell itself at the time of colony formation.