Fungal pathogens have plagued the Pyropia species of red algae (Rhodophyta) and seriously reduced the output of the Pyropia aquaculture industry in Korea and Japan (Kawamura et al. 2005, Kim et al. 2014). Red rot disease is a major algal disease that was first reported in Porphyra tenera (= Pyropia tenera) from Japan (Arasaki 1947). After that, Pythium species, the etiological agent of red rot disease, has been isolated and characterized from the red rot of infected Pyropia species (Takahashi et al. 1977).
Among species of the genus Pythium, P. chondricola, P. porphyrae, and P. adhaerens are closely related, based on the characteristics of a filamentous non-inflated sporangia, slow growth, and 1-4 diclinous antheridia (Levesque and De Cock 2004). However, these species show different host / substrate-specific relationships (Matsumoto et al. 1999, Levesque and De Cock 2004). Pythium porphyrae is recognized as the only pathogen of red rot disease in Pyropia species (Takahashi et al. 1977, Kim et al. 2014), while P. adhaerens has been isolated from soil and P. chondricola was discovered in Chondrus crispus (Levesque and De Cock 2004).
To verify the taxonomy of the genus Pythium, Matsumoto et al. (1999) analyzed the nuclear internal transcribed spacer (ITS) sequences of 30 Pythium species. More recently, Levesque and De Cock (2004) examined the phylogenetic relationship among 102 isolates using sequences of the ITS and D1, D2, and D3 regions in nuclear ribosomal DNA of Pythium. In addition, a DNA barcoding study for Oomycetes was conducted using ITS and cytochrome c oxidase subunit 1 (cox1) markers, and the taxonomic resolution of these two molecular markers was compared (Robideau et al. 2011).
Recently, Schroeder et al. (2013) suggested primers for polymerase chain reaction (PCR) based methods for the diagnosis and quantification of Pythium species. In a molecular phylogenetic tree from the nuclear ribosomal DNA and cox1 region of Pythium, P. adhaerens, P. chondricola, and P. porphyrae formed a single clade (Levesque and De Cock 2004, Robideau et al. 2011). In another approach, Park et al. (2001, 2006) developed a P. porphyrae specific ITS marker and applied it to forecasting red rot disease in Pyropia yezoensis cultivation farms.
Due to the increasing economic significance of Pyropia species, concern for algal diseases in the scientific community and aquaculture industry has been raised (Gachon et al. 2010). Therefore, accurate species identification of the algal pathogen is needed to clarify the host-parasite relationship and to help find a scientific solution for the disease. Recent fungal DNA barcoding study has revealed that the ITS region cannot provide sufficient taxonomic information to discriminate between the closely related Pythium species (Robideau et al. 2011). This study has shown the taxonomic usefulness of the cox1 region at the species level and suggested it as another candidate molecular marker for DNA barcoding. In this study, we applied the ITS and cox1 markers to identify Korean Pythium species from an infected blade of Pyropia yezoensis, from a Pythium culture strain, and from environmental seawater. Using Pythium specific ITS primers and newly designed cox1 primers, we accurately identified the species of Pythium and revealed its taxonomic relationship among other Pythium species.
We sampled a blade of Pyropia yezoensis infected by red rot disease in the field (Dec 2014 , Biando, Gunsan, Korea). A preliminary determination of infection by an algal pathogen was conducted based on the morphological characteristics in the diseased plant. Another Pythium strain was also successfully isolated from an infected Pyropia yezoensis specimen collected from the Korean coast (Dec 2014, Aphaedo, Shinan, Korea) and cultured under indoor conditions using cornmeal agar (Park et al. 2001). This culture strain has been deposited in the Seaweed Research Center (National Fisheries Research and Development Institute, Mokpo, Korea). We also analyzed DNA templates from previous metagenomic studies of seawater (Lee et al. 2010, 2012, Yoon et al. 2012) and from environmental seawater sampled from the Nakdong River estuary near the Pyropia aquaculture region (Aug 5, 2011, Korea).
We applied molecular markers from the ITS and cox1 regions for this study. For the amplification of the ITS region, we used Pythium porphyrae specific ITS primers (PP-1/PP-2) (Park et al. 2001). To target the cox1 region of Pythium species, we designed cox1 primers having putative specificity for the genus Pythium. For the design of these cox1 primers, we downloaded the complete cox1 sequence of P. ultimum (NC_014280) and compared it with other oomycete cox1 sequences through the BLAST searching option in GenBank (National Center for Biotechnology Information, NCBI). From the cox1 sequence alignment, we found a conserved cox1 region among Pythium species. The new cox1 primers for the genus Pythium were designed basing on this conserved region.
The DNA extraction, PCR, and sequencing were conducted following methods outlined in Lee et al. (2011). Total genomic DNA was extracted from the blade of infected Pyropia yezoensis using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. We also extracted total genomic DNA from environmental seawater using protocols suggested by Lee et al. (2010). Amplifications were carried out in a final volume of 20 μL using amfiXpand (GenDEPOT, Barker, TX, USA) with PCR conditions of 2 min at 95℃, 40 cycles of 30 s at 94℃, 30 s at 48℃, and 1 min at 72℃, with a final 7 min extension step at 72℃.
PCR products were subsequently sequenced in both directions by commercial sequencing (Genotech, Daejeon, Korea) and chromatograms were analyzed using Sequencher 5.3 (Gene Codes Corporation, Ann Arbor, MI, USA). The sequence similarity analyses of ITS and cox1 sequences were conducted BLAST in the GenBank (NCBI). For the phylogenetic analyses, the ITS and cox1 reference sequences of related Pythium species were obtained from GenBank, and all sequences were aligned using Clustal X v.1.8 (Thompson et al. 1997). A phylogenetic tree was constructed by the neighbor-joining method (NJ) (Saitou and Nei 1987) using PAUP 4.0 (Swofford 2001). A bootstrap analysis with 2,000 replicates was conducted to assess the robustness of the NJ tree.
In addition to the previously developed ITS primers (Park et al. 2001), new PCR primers were designed for the selective amplification of the cox1 region of Pythium species, since the diverse fungus can be present in decaying seaweed (e.g., the infected Pyropia blade) and environmental seawater samples. The forward primer (cox1-pyth-F1; 5′-ATTAGAATGGAATTAGCACAAC-3′) is positioned at 36405-36426 of the mtDNA of P. ultimum (NC_014280) and the reverse primer (cox1-pyth-R1; 5′-CTTAAACCWGGAGCTCTCAT- 3′) is bound at position 36813-36832.
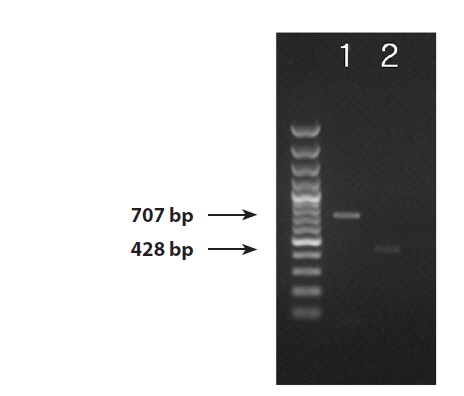
We successfully obtained PCR bands from the DNA extractions from the infected blade of Pyropia yezoensis, thePythium culture strain, and the environmental seawater samples. PCR products 707 bp and 428 bp in size were amplified by the ITS and cox1 primers (Fig. 1). The ITS region was successfully amplified from all three sample sources (the blade of Pyropia, Pythium culture strain, and environmental seawater). Even though the amplification yielded bands that were very weak from the ITS region of the sample from environmental seawater, the sequence was successfully determined. On the other hand, we could amplify the cox1 region only from the blade of Pyropia and the culture strain.
From the similarity analysis using BLAST, the ITS sequences showed no variation among Korean isolates (the blade of Pyropia, Pythium culture strain, and seawater). In addition, the Korean ITS sequences had 100% sequence similarity with the reference sequences of ITS in P. porphyrae (Korea, AB043506; Japan, AY598673, AB185111, AB043506; USA, JQ898472) and P. chondricola (Netherlands, AY598620, HQ643496, HQ643497, HQ643498; USA, HQ643499). The Korean cox1 sequences in this study showed 100% similarity with P. chondricola (Netherlands, HQ708542, HQ708543, HQ708544; USA, HQ708545), 99% (382/386; matched sequences / total sequences excluding the primer binding sites) with P. porphyrae (Japan-HQ708794) and 99% (381/386) with P. adhaerens (HQ708462).
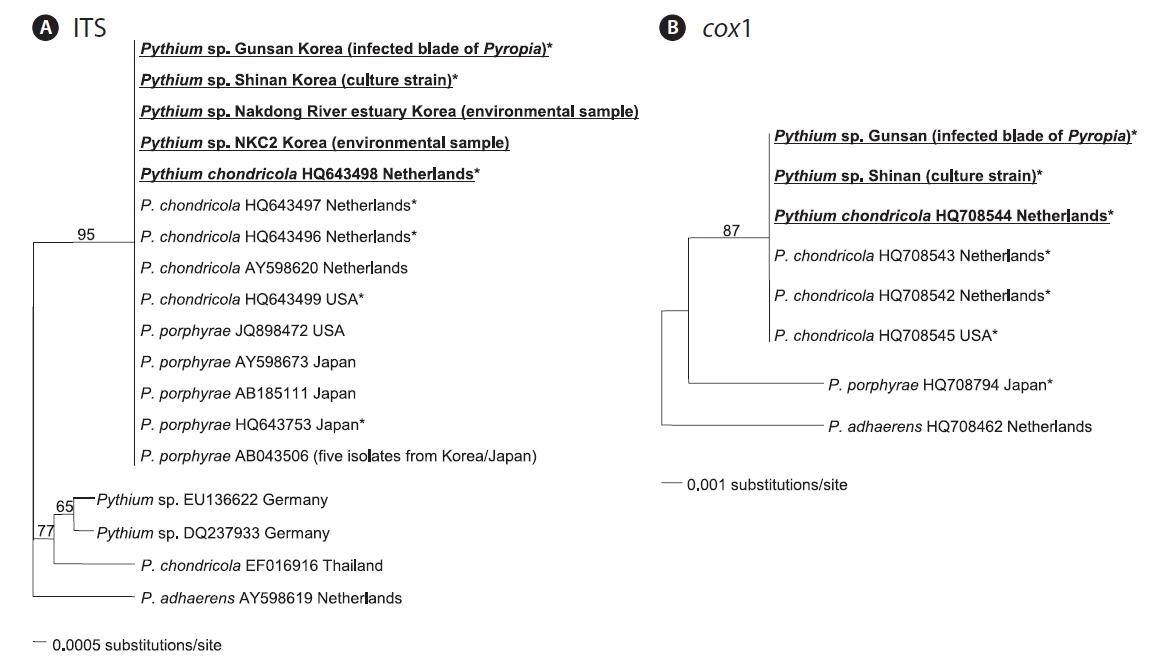
In the ITS phylogenetic tree, the Korean Pythium ITS sequences clustered in a single clade with P. porphyrae (Korea, Japan, and USA) and P. chondricola (Netherlands), and also with a more distantly related P. chondricola (EF016916) from Thailand (Fig. 2A). In contrast, the cox1 phylogenetic tree showed that Korean cox1 sequences formed a single clade only with P. chondricola cox1 sequences reported from the Netherlands and USA (Fig. 2B). Pythium porphyrae reported from Japan formed a distant sister group of the clade including Korean Pythium species and P. chondricola (Netherlands and USA).
Korean Pythium isolates (in this study) had the same ITS sequence as P. porphyrae and P. chondricola sequences that were previously reported. Therefore, the ITS sequence did not seem to provide sufficient information to resolve the taxonomic relationship between P. porphyrae and P. chondricola. The only difference in the ITS sequences of P. porphyrae / P. chondricola was in the poly A sequence at the 5′ end of the ITS region (6 in HQ643499 vs. 7 in other isolates). On the other hand, the cox1 sequence showed sufficient variation to discriminate between P. porphyrae (HQ708794) and its close relatives, P. chondricola (HQ708545) and P. adhaerens (HQ708462) (Fig. 2).
A recent DNA barcoding study also reported the greater discriminative power in the cox1 region than in the ITS region, allowing a clarification of the taxonomic relationships among the closely related Pythium species (Robideau et al. 2011). From our results, the molecular analysis based on the ITS region could not provide specific information for the identification of P. porphyrae. Therefore, the ITS sequence was not a suitable barcoding marker to discriminate P. chondricola from P. porphyrae.
From the PCR analysis of the environmental seawater samples using the Pythium specific ITS primer set, we detected P. chondricola / P. porphyrae ITS sequences from the coastal seawater of Korea. We did not obtain a cox1 amplicon from these samples. Because nuclear ribosomal DNA has a high number of tandem repeat sequences (Rogers and Bendich 1987), PCR amplification with ITS primers could be more efficient than cox1 region in detecting Pythium.
The ITS sequences from environmental samples could have originated from fungal zoospores of P. chondricola / P. porphyrae in the seawater. Kawamura et al. (2005) also reported the distribution of P. porphyrae in the seafloor sediment (Ariake Sea, Japan) basing on an ITS sequence analysis. Therefore, P. chondricola / P. porphyrae appears to be present in the Korean aquatic ecosystem and could infect Pyropia species. This study provides important information to monitor the distribution of red rot disease. In addition, more sensitive, high efficiency methods should be developed to detect Pythium species from environmental seawater samples because relatively little biomass of the zoospore exists in the aquatic ecosystem.
Pythium chondricola was first isolated from decaying Chondrus crispus collected in and near the salt lake Grevelingen in the Netherlands (De Cock 1986). Since it was as a new species, P. chondricola has only been reported from the Netherlands and USA using molecular markers (Robideau et al. 2011). De Cock (1986) examined different fungal isolates from the same origin and locality as earlier isolates of P. chondricola, and discovered diverse sources of P. chondricola, including aquatic plants such as Zostera marina (flowering plants), Ulva lactuca (green algae), and an unidentified red alga. This implies that P. chondricola can infect a variety of host plants and that Pyropia species is one of them.
Robideau et al. (2011) reported the presence of P. porphyrae (HQ708794) from Japan as a separate taxonomic entity from P. chondricola. Pythium chondricola has not been reported from Pyropia species or aquatic environments of Korea or Japan (Kawamura et al. 2005, Park et al. 2006, Uzuhashi et al. 2015). To our knowledge, the finding of P. chondricola in this study is the first report of this species in the Asian region (Global Catalogue of Microorganisms [GCM]; The Barcode of Life Data System [BOLD]) (Ratnasingham and Hebert 2007, Wu et al. 2013).
We could hardly find P. porphyrae from Pyropia blades during this study. Therefore, further studies targeting the cox1 region are strongly requested and should include the wide survey for red rot disease in Pyropia cultivation sites and in the natural aquatic ecosystem to confirm whether P. chondricola and P. porphyrae are coexisting as algal pathogens of Pyropia species in Korea.