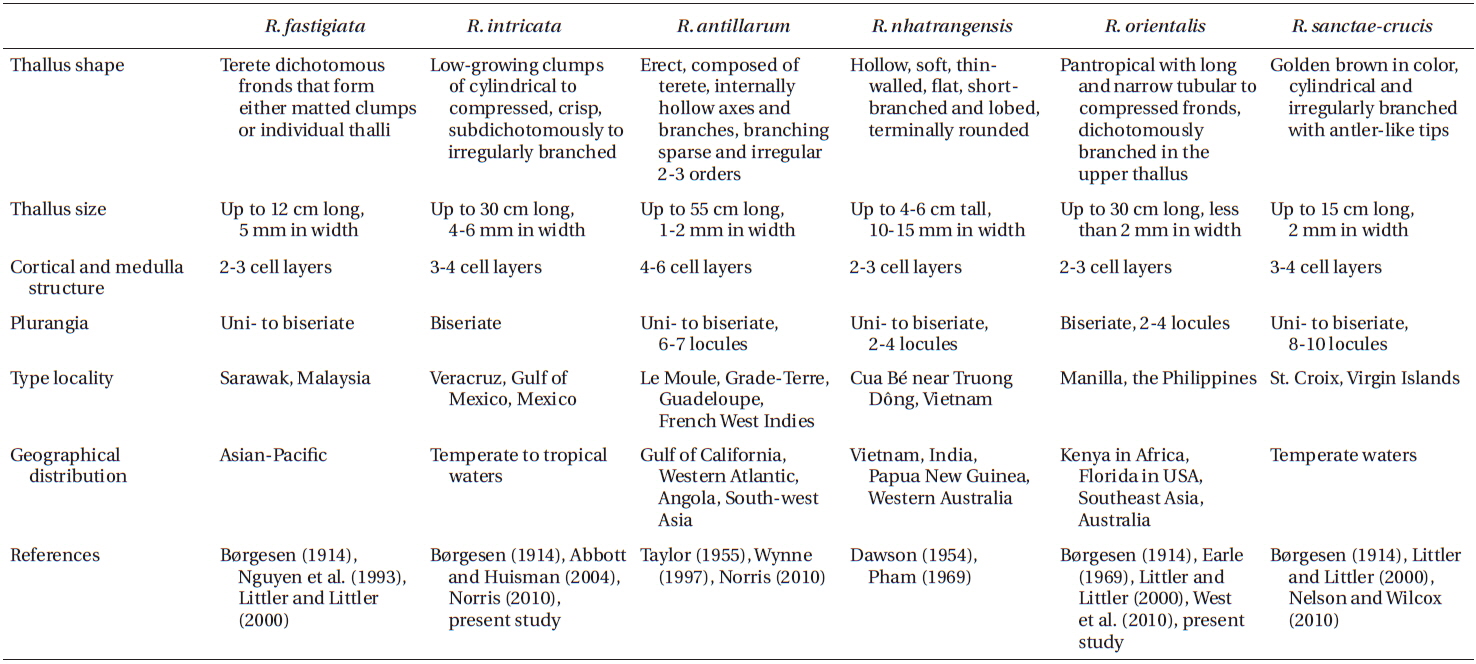
The genus Rosenvingea Børgesen is a tropical to subtropical genus of scytosiphonacean brown algae that includes seven currently accepted species (Guiry and Guiry 2014). Rosenvingea is distinguished by its erect thalli with alternate or dichotomous branches, an apical to subapical meristem, a single plastid and prominent pyrenoid in vegetative cells, hollow medulla, and phaeophycean hairs and plurangial sori scattered on the thallus surface (Børgesen 1914, Abbott and Huisman 2004, Norris 2010, West et al. 2010). Børgesen (1914) established the genus with R. sanctae-crucis Børgesen as the type and included three other species in the genus Asperococcus J. V. Lamouroux as follows, R. fastigiata (Zanardini) Børgesen, R. intricata (J. Agardh) Børgesen, and R. orientalis (J. Agardh) Børgesen. Subsequently three more species were added: R. antillarum (P. Crouan & H. Crouan) M. J. Wynne, R. floridana (W. R. Taylor) W. R. Taylor, and R. nhatrangensis E. Y. Dawson (Dawson 1954, Taylor 1955, Wynne 1997). However, the species circumscription in Rosenvingea is very confused. For example, R. sanctae-crucis, the type of Rosenvingea, was suggested to be conspecific with R. orientalis (Earle 1969, Littler and Littler 2000, Wynne 2005, Dawes and Mathieson 2008, Nelson and Wilcox 2010). Wynne (1997) treated R. floridana as a synonym of R. antillarum, although Norris (2010) considered that both species are not conspecific. The cosmopolitan or endemic distribution patterns of Rosenvingea species are, accordingly, doubtful.
The first molecular analysis of Rosenvingea was based on sequences of rbcL and rbcS from Japanese specimen of R. intricata (Kogame et al. 1999) and was followed with psaA by Cho et al. (2006). Subsequently, R. intricata was analyzed with additional seven genes, plastid atpB, psaA, and psbA, and mitochondrial atp9, cox1, cox3, and nad11 (Silberfeld et al. 2010). The second species included in molecular taxonomic study is R. orientalis that was sequenced for psaA and cox3 (West et al. 2010, Lee et al. 2014).
Dawson (1954) described R. nhatrangensis as a new species and also reported R. orientalis in Nha Trang, Vietnam. Afterwards, R. intricata was reported in southern Vietnam (Pham 1969) and additional record from northern Vietnam (Nguyen et al. 1993). Another species, R. fastigiata was also reported to occur in northern Vietnam (Nguyen et al. 1993, 2013). We collected two species of Rosenvingea in Nha Trang during March and April 2011. Together with morphological observations, we analyzed both cox3 and psaA gene from these specimens so as to provide a better understanding of species diversity and phylogenetic relationships in Rosenvingea.
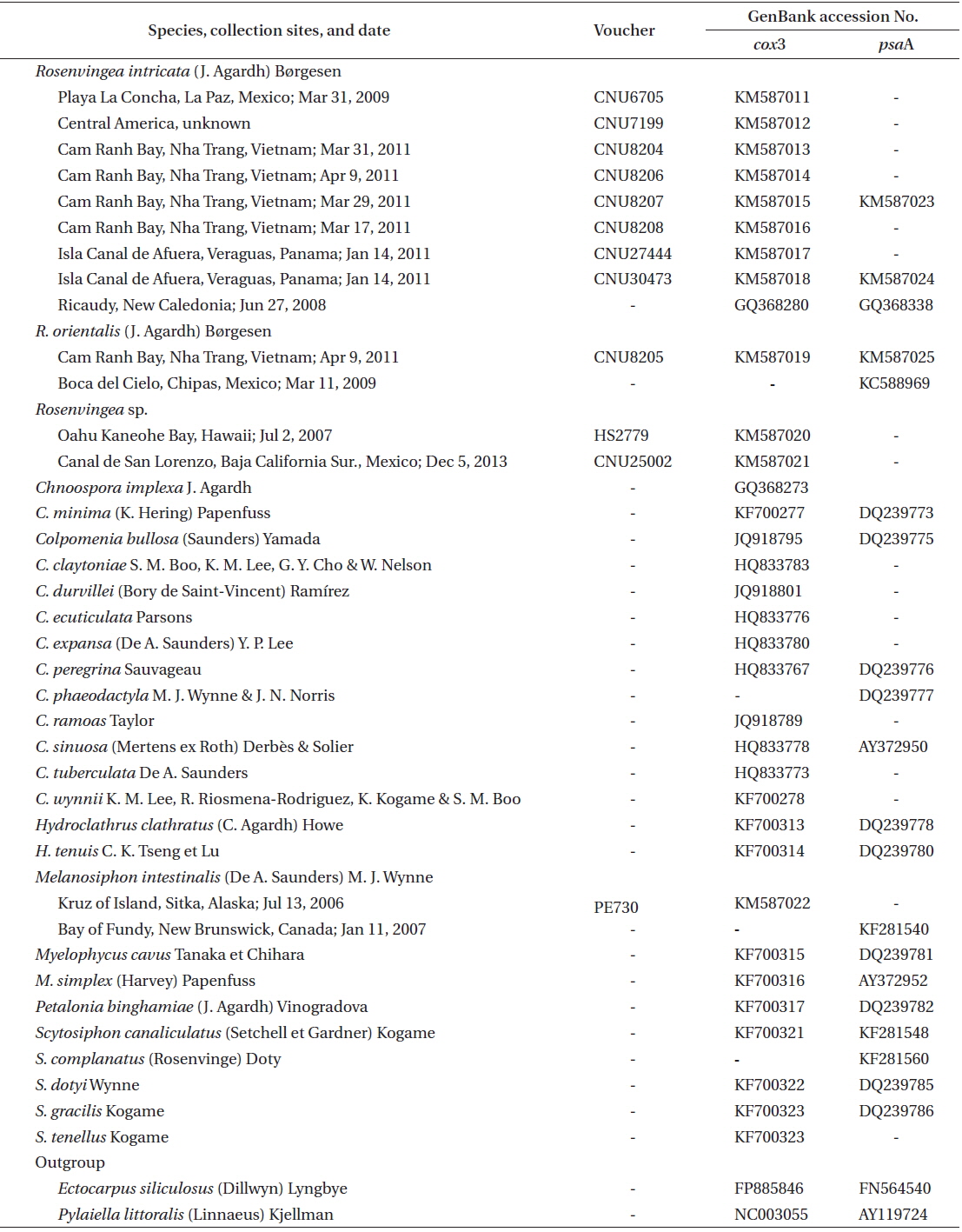
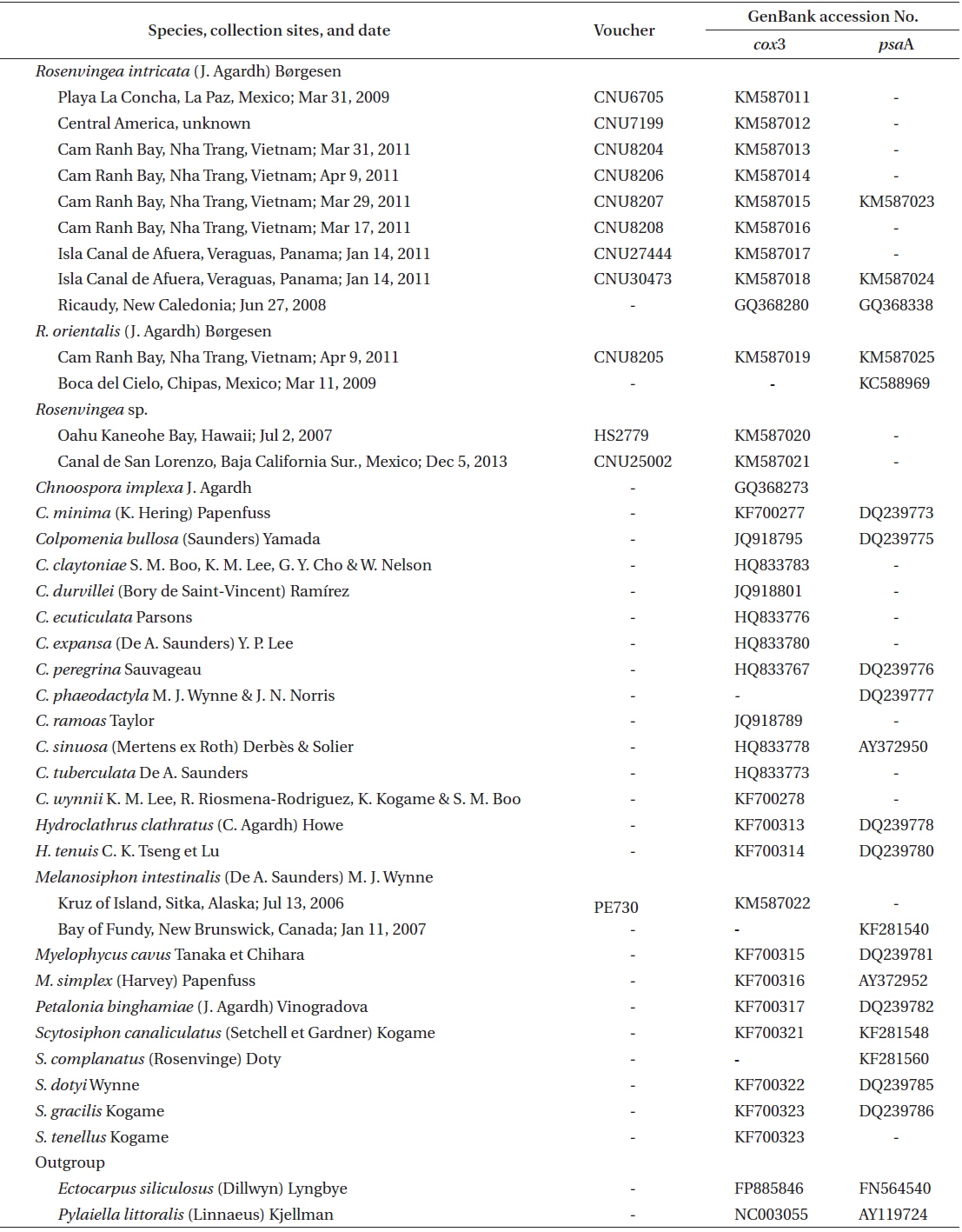
Representative Rosenvingea specimens were collected from mid intertidal zone in Cam Ranh Bay, Nha Trang, Vietnam, and specimens collected in Mexico and Panama were also included in analyses for comparison (Table 1). All specimens collected were mounted on herbarium sheets. Small parts of the specimens were cut for molecular analyses. Photographs were taken with a DP-71 camera (Olympus, Tokyo, Japan) attached to a BX-51 microscope (Olympus). Voucher specimens were deposited at the Department of Biology, Chungnam National University, Daejeon, Korea (CNUK).
Information on specimens used for the molecular study is given in Table 1. DNA extraction, PCR amplification, and sequencing followed Boo et al. (2011b). Primer pairs for the amplification and sequencing of each gene were F49–R20 for cox3 (Boo et al. 2010, 2011a) and psaA130F-psaA940R and psaA870F-psaA1760R for psaA (Yoon et al. 2002).
Phylogenies of cox3 and psaA datasets were reconstructed using maximum likelihood (ML), including Ectocarpus siliculosus (Dillwyn) Lyngbye and Pylaiella littoralis (Linnaeus) Kjellman as outgroups. ML analyses were performed with RAxML v.7.2.8 (Stamatakis 2006) using the GTR + GAMMA + I model. We used 200 independent tree inferences, applying options of automatically optimized subtree pruning regrafting rearrangement and 25 distinct rate categories in the program to identify the best tree. Statistical support for each branch was obtained from 1,000 bootstrap replications with the same substitution model.
Bayesian inference was performed for individual datasets with MrBayes v.3.2.1 (Ronquist et al. 2012) using the Metropolis-coupled Markov Chain Monte Carlo (MC3) with the GTR + GAMMA + I model. For each matrix, two million generations of two independent runs were performed with four chains and sampling trees every 100 generations. The burn-in period was identified graphically by tracking the likelihoods at each generation to determine whether they reached a plateau. The 40,002 trees sampled at the stationary state were used to infer Bayesian posterior probabilities (BPP).
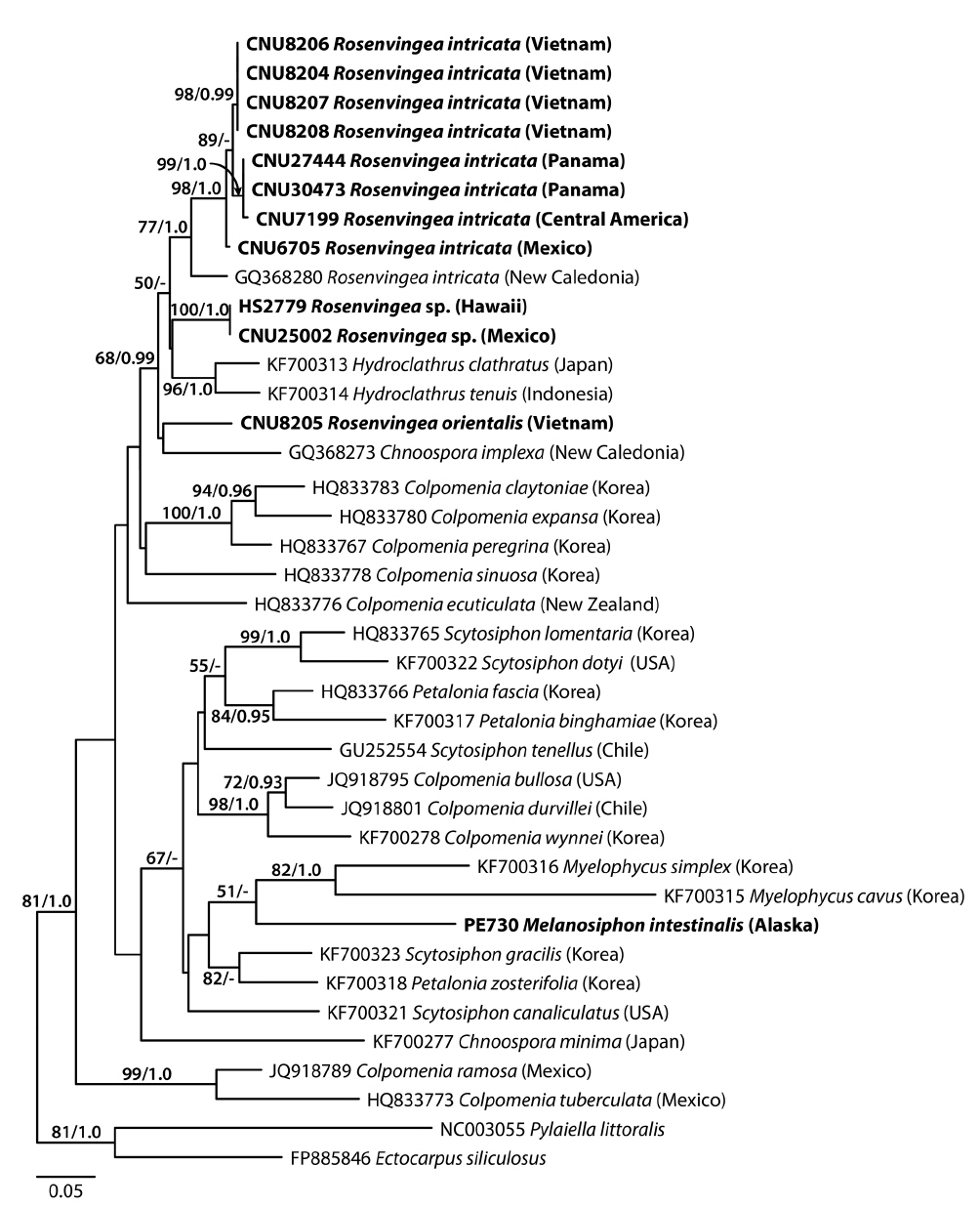
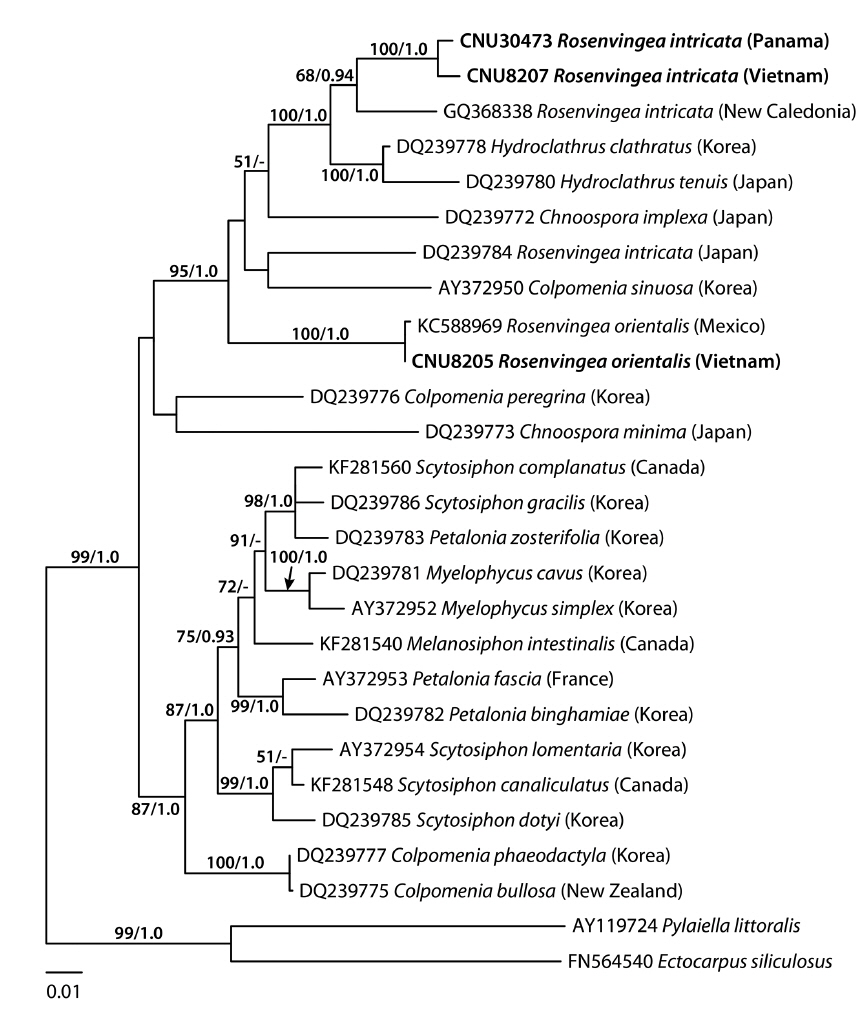
A 627-nucleotide portion of cox3 was aligned for 40 sequences including 12 Rosenvingea and one previously published sequence of R. intricata from New Caledonia. Variable sites occurred at 284 positions (45.3%), of which 222 positions (35.4%) were parsimoniously informative. R. intricata differed by 32-39 bp (5.5-6.7%) from Rosenvingea sp. collected in Hawaii and Mexico and by 40-45 bp (6.9-7.7%) from R. orientalis from Vietnam. There was a difference of 42 bp (7.2%) between Rosenvingea sp. and R. orientalis. One sequence from New Caledonia (GQ368280) was highly divergent from other sequences of R. intricata (28 bp, 4.8%). Pairwise divergence within R. intricata (except the New Caledonia specimen) ranged up to 8 bp (1.4%). The phylogenetic relationships of Rosenvingea, using cox3 sequences, showed a well-supported Scytosiphonaceae consisting of eight genera (Fig. 1). There was no resolution of the phylogenetic relationships among sequences of R. intricata sequences.
A 1,488-nucleotide portion of the psaA gene was compared across 28 sequences including six Rosenvingea and three previously published sequences collected from New Caledonia, Mexico and Japan. Variable sites occurred at 392 positions (26.3%), of which 256 positions (17.2%) were parsimoniously informative. R. intricata differed by 88-98 bp (6.3-7.0%) from R. orientalis. The psaA sequence of R. intricata from the Vietnamese specimen differed by 11 bp (0.8%) from the specimen from Panama, 54 bp (3.9%) from the New Caledonian specimen, and 92 bp (6.6%) from the Japanese specimen. In the psaA tree (Fig. 2), R. intricata from Vietnam and Panama together with R. intricata from New Caledonia formed a monophyletic group with moderate support values (68% for ML and 0.94 for BPP). This group was closely related to Hydroclathrus with maximum support values. One published sequence of R. intricata collected from Japan did not belong to the group of samples from Vietnam and Panama, but rather sister to Colpomenia sinuosa (Mertens ex Roth) Derbès & Solier. R. orientalis was placed in the clade that includes R. intricata, Hydroclathrus clathratus (C. Agardh) Howe, H. tenuis C. K. Tseng & Lu, Chnoospora implexa J. Agardh, and Colpomenia sinuosa (95% for ML and 1.0 for BPP).
Thalli of R. intricata were brown to dark brown in color, up to 5 cm in height and 5 mm in diameter (Fig. 3A-D). The thalli formed a dense decumbent mat with many branches that were often inter-adhesive. The axes were cylindrical to compressed, subdichotomously to irregularly lobed, 4-6 mm in diameter, becoming narrower distally. Apices of branches were mostly rounded.
Thalli of R. orientalis were up to 18 cm long with narrow (less than 2 mm in width), tubular to compressed fronds that are dichotomously branched in the upper thallus (Fig. 3E-H). The thalli were golden to light brown in color, with hollow axes, up to 2 mm in diameter. Branches were dichotomous to irregularly lobed, usually tapering at the base and apex, ends of branches nearly hair-like.
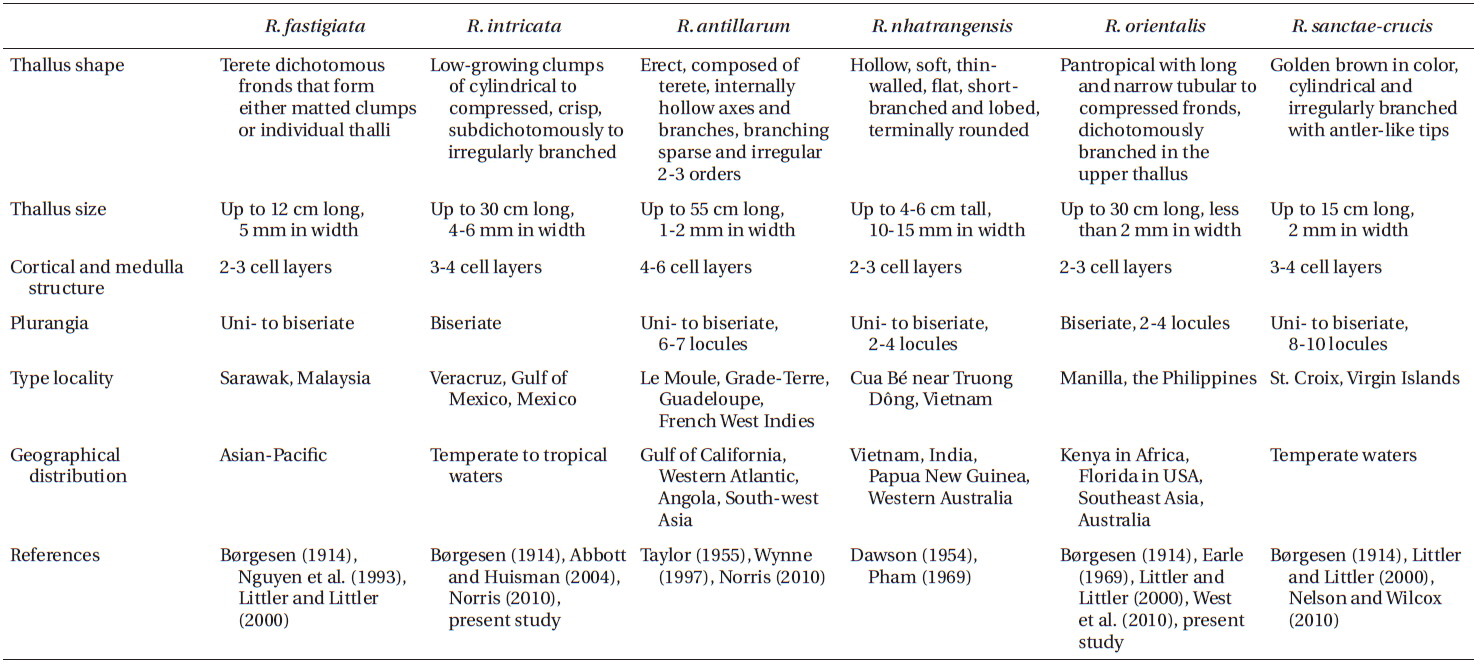
Our analyses of mitochondrial cox3 and plastid psaA sequences clearly showed the occurrence of two genetically and morphologically distinct species of Rosenvingea from Nha Trang, Vietnam. We collected both species in Cam Ranh Bay, Nha Trang, Vietnam, and have assigned these species to R. intricata and R. orientalis. Morphological comparison with other Rosenvingea species is given in Table 2.
R. intricata is the most common species in the genus reported in tropical to subtropical waters of the world (see Guiry and Guiry 2014). In cox3 and psaA trees (excluding the Japanese taxon in the cox3 tree due to the unavailability of the samples), R. intricata was separated into three different taxa. The first group was composed of specimens from Vietnam, Mexico, and Panama. The morphology of our plants is consistent with the description and illustration of R. intricata (Børgesen 1914, Earle 1969). We therefore recognized this taxon as R. intricata because the clade in our analyses included specimens collected from near the type locality in Veracruz, Mexico. Accordingly, our study confirmed the previous reports on the occurrence of R. intricata in Vietnam (Pham 1969, Nguyen et al. 1993) as well as Baja California in Mexico (Norris 2010) and Panama (Wysor and De Clerck 2003). The second taxon was the New Caledonia taxon (as GQ368280 in cox3, GQ368338 in psaA) that formed a sister to R. intricata. The pairwise divergence of psaA (54-55 bp, 3.9%) between the New Caledonian taxon and R. intricata is similar or higher than that between scytosiphonacean species (e.g., 2.2% between Scytosiphon lomentaria and S. dotyi) (Cho et al. 2006). The third taxon distinguished consisted of the Japanese specimens (DQ239784) and was distantly related to other taxa of Rosenvingea. These results reveal an urgent need for further molecular analysis of specimens currently assigned to R. intricata in various countries (see Guiry and Guiry 2014).
R. orientalis is distributed in subtropical to tropical waters of Kenya, Florida in USA, Southeast Asia, and Australasia (see Guiry and Guiry 2014). Its type locality is Manilla, the Philippines, and its diagnostic characters are the tubular to compressed thalli, the tall size (up to 30 cm), narrow diameter of branches (less than 2 mm), and dichotomous branches (Børgesen 1914, Earle 1969, West et al. 2010). Following previous phycologists’ opinions (Earle 1969, Littler and Littler 2000, Abbott and Huisman 2004, Wynne 2005), Nelson and Wilcox (2010) reported that R. orientalis may be conspecific with R. santae-crucis and the former has priority. Molecular analysis of type specimens or fresh collections of R. santae-crucis near the type locality are needed to resolve about the conspecificity of these taxa.
The absence of R. nhatrangensis in our collections despite our repeated samplings in the type locality, Nha Trang, Vietnam, suggests that there is an urgent need for an analysis of the type specimens of this and related taxa. R. nhatrangensis is distinguished by much inflated, lobed and short thalli with its wide diameter of branches (2.5-3.0 cm in lower part of erect thallus and 1.0-1.5 cm in the branches) (Dawson 1954). R. fastigiata var. major Reinbold reaches a branch diameter of 5 mm, and bears sori in rings around the hair groups. However, based on figures and description by Dawson (1954), Egerod (1974) suggested that R. nhatrangensis may be conspecific with R. fastigiata var. major. Kochang Archipelago, Thailand, the type locality of this variety is close to Nha Trang, Vietnam, the type locality of R. nhatrangensis (Reinbold 1901, Dawson 1954). Fresh collection of R. fastigiata var. major in the type locality is necessary to confirm their conspecificity.
This is the first study using mitochondrial cox3 sequences to reveal the non-monophyly of the genus Rosenvingea. Our analyses showed that Rosenvingea that clustered with Hydroclathrus ssp., and Chnoospora implexa. This topology was also reflected in the psaA tree with the inclusion of Colpomenia sinuosa. The nonmonophyly of Rosenvingea revealed in the present study indicates the need for a reconsideration of the generic concept. However, based on the cox3 and psaA trees, it is to date impossible to combine or rearrange these genera based on molecular data as none of the genera are monophyletic in the analyses. Chnoospora, Colpomenia, Petalonia, and Scytosiphon in the family Scytosiphonaceae have been confirmed to be non-monophyletic in rbcL and / or psaA, and cox3 phylogenies (Kogame et al. 1999, Cho et al. 2006, Lee et al. 2014). As discussed by Kogame et al. (1999) and Cho et al. (2006), further investigations based on taxon-wide sampling, and employing other supplementary gene sequences, life histories, and ultrastructure of gametes, are urgently needed to clarify the relationships of all these currently non-monophyletic genera of the Scytosiphonaceae.
In conclusion, we confirmed the occurrence of R. intricata and R. orientalis from Nha Trang, Vietnam. Our study therefore reveals that both species are distributed from Vietnam to Mexico and / or Panama. Further collection and careful examination of specimens will probably extend the known distributions of both species to surrounding Vietnamese waters. Additional investigations are needed to confirm the relationships of R. nhatrangensis with R. intricata and R. fastigiata var. major. The finding of cryptic species within R. intricata highlights the fact that the number of Rosenvingea species in the world inventory will increase with additional collections.