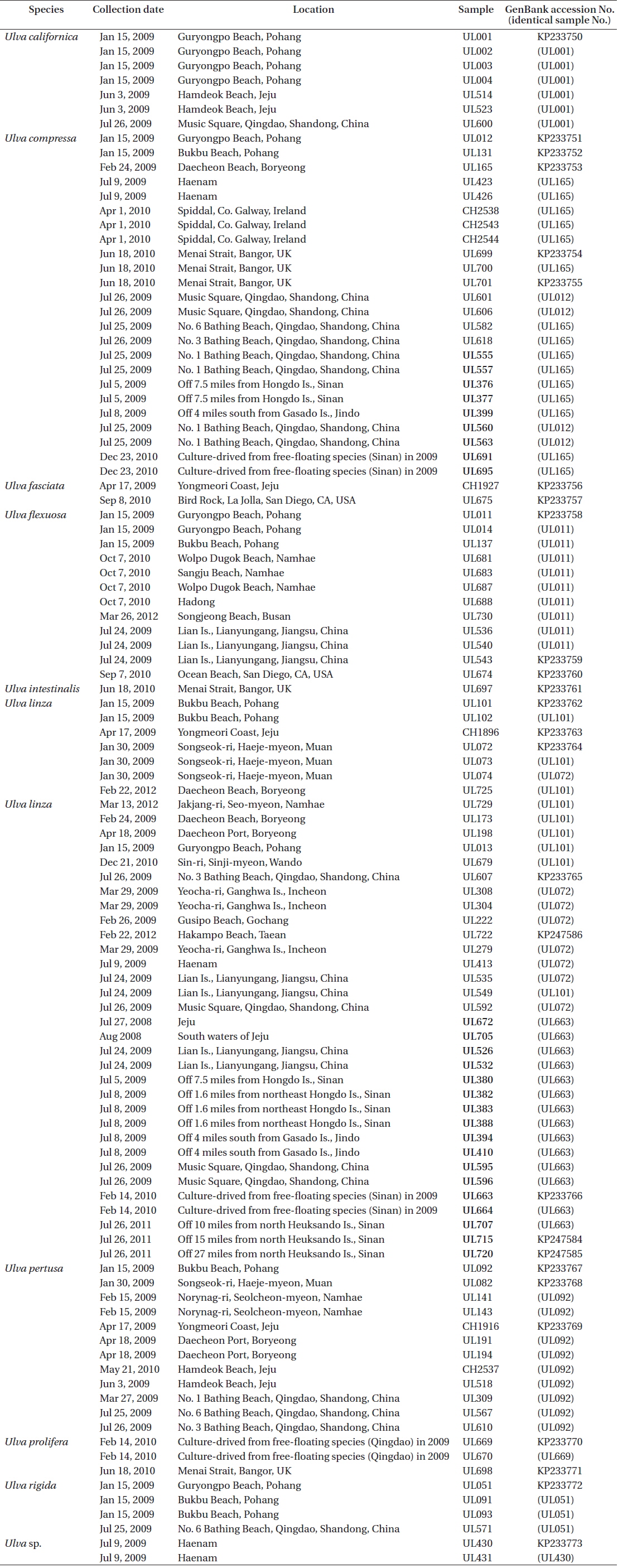
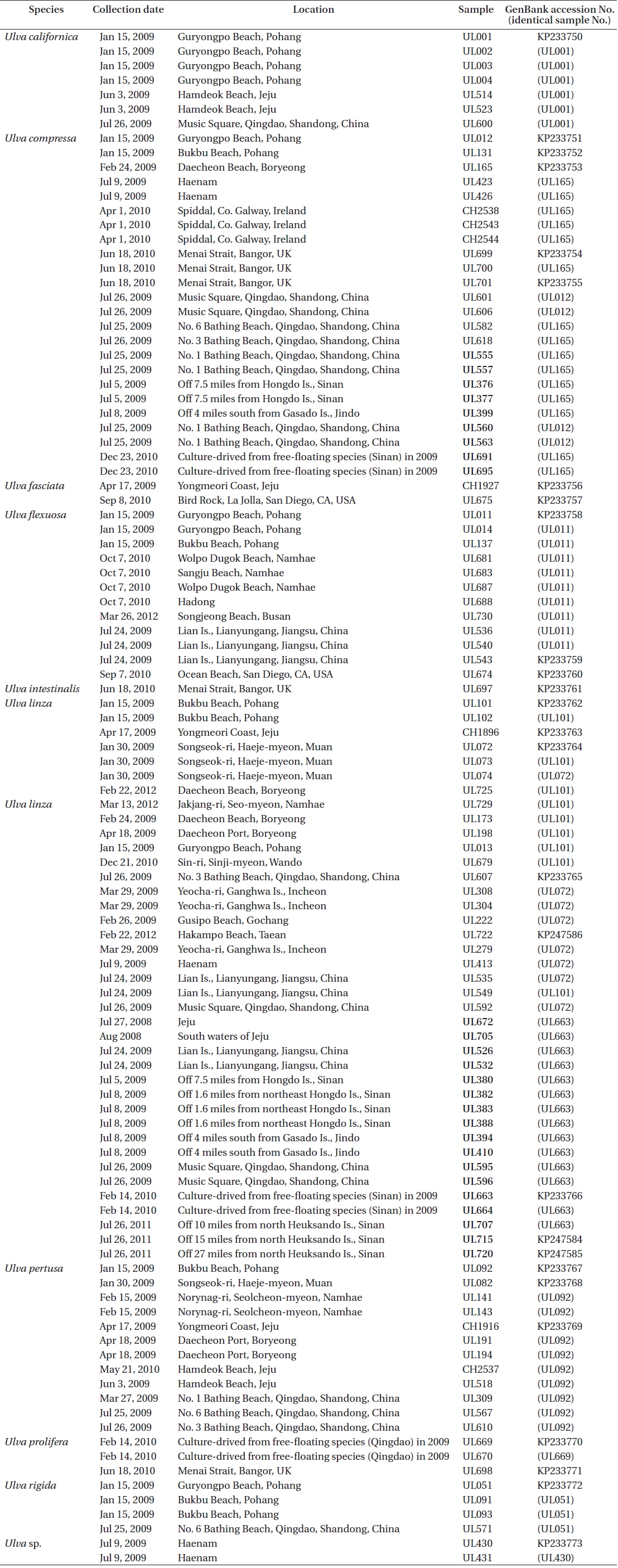
Macroalgal blooms occur worldwide, but the green tide that occurred on the Qingdao coast in late June 2008 was unprecedented, as it formed the largest green algal bloom recorded. Massive floating green mats covered about 1,200 km2 of coastal water, and the Qingdao shoreline on the northeastern coast of China. This phenomenon reoccurred in the same region in July 2009, covering much of the same area as the previous year (Zhang et al. 2011). It has subsequently become an annual phenomenon.
The origin of the floating green mats off the east coast of China from 2007 to 2009 has been studied extensively, and different explanations have been a source of considerable controversy. Liu et al. (2009, 2010a) suggested that the floating green mats originated from Porphyra cultivation systems, whereas Pang et al. (2010) hypothesized a land-based pond origin associated with animal aquaculture. Despite these differences in opinion, the fundamental cause of these green algal blooms was eutrophication. Accordingly, a consensus was reached that the green tides in the Yellow Sea (YS) are caused by high nutrient conditions.
Green tide-forming species occur in two forms, such as attached thalli or free-floating thalli. These forms are based on substratum availability and environmental conditions. Ulva spp. have high intraspecific morphological plasticity associated with seasonal and environmental conditions (Blomster et al. 1998, 1999). This plasticity reflects competition in complex and varying environments where survival depends on adaptation to limited nutrient, light, salinity, and temperature conditions. A consequence of the morphological plasticity is that some Ulva spp. (e.g., Ulva compressa and U. intestinalis) cannot be identified using morphological characters, e.g., changes in branching induced by low salinity or salinity shock (Blomster et al. 1998). Furthermore, laboratory culture experiments show that bacteria and their exudates associated with Ulva may induce extensive morphological changes that cross supposedly well-defined species boundaries (Provasoli and Pintner 1980, Marshall et al. 2006). Thus, identifying Ulva spp. based on morphological characters can be problematic. For this reason, the Ulva sp. that appeared in the YS was not identified correctly, and some researchers attempted to identify the species based on molecular phylogenetic characters.
A molecular phylogenetic evaluation of Ulva isolated from the 2008 Qingdao bloom also showed conflicting results. Leliaert et al. (2009) concluded that the species from 2008 Qingdao was part of the Ulva linza-procera-prolifera (LPP) complex, whereas Wang et al. (2010) concluded that the alga was U. prolifera. Zhang et al. (2011) reported that the blooming species forming the floating mats in the YS in 2008 and 2009 was U. prolifera based on a 5S rDNA spacer sequence analysis.
Following the Qingdao bloom a similar event, albeit on a smaller scale, occurred near the Jeju Island coast in 2008. Green macroalgal patches appeared to the west of Ieodo Ocean Research Station and were 100-200 m wide and 200-800 m long in 2008 (Choi et al. 2010). The same phenomenon occurred in the southwest sea of Korea (the southeastern part of the YS) in 2009, and the green mats were senescent with reduced physiological activity (Kim et al. 2011). The green tide reoccurred in the southwest sea of Korea in July 2011, and off the coasts of China (the western part of the YS), and this has become an annual event in the YS.
Green tides in the southwest sea of Korea were similar to the Chinese blooms, but have not been studied to the same extent as those in China. Nevertheless, satellite images from the southeastern part of the YS suggest that the green patches on the west coast of Korea originated from the east coast of China (Son et al. 2015). It is clear that the green tides in the YS have been reoccurring every year since 2007. A key step to understand the reason behind this nuisance phenomenon and to potentially manage it is to identify the species involved. In this study, we attempted to identify the bloom-forming species off the Korean coast using both morphological approaches and a phylogenetic analysis of plastid ribulose-1,5-bisphosphate carboxylase (rbcL) gene sequences. In addition, we re-evaluated the identity of the YS green tide species.
Over 700 specimens of Ulva spp. were collected on and off the coasts of Korea and China including bloom-forming species in the YS from 2008 to 2012 (Table 1). The floating and benthic Ulva samples from the Chinese coasts were collected in the summer of 2009 when floating algal mats appeared on the southwestern sea of Korea. The samples were rinsed in deionized water, conspicuous epiphytes were removed, and the samples were preserved in silica gel until DNA extraction. Voucher specimens were prepared as herbarium specimens and deposited at Chonnam National University.
Morphological characters, such as cell shape, size, and arrangement, as well as frond margin shape and number of pyrenoids, were determined by light microscopy. Most samples were examined from fresh material and dried samples were observed after rehydration in seawater.
Genomic DNA from silica-dried samples was ground with a mortar and pestle in liquid nitrogen and extracted using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The plastid-encoded rbcL gene was amplified by the polymerase chain reaction (PCR) using the RH1, 1385r, SHF1, SHR4, rbc590, rbc571, and KF2 primers (Table 2) and a general PCR reaction mixture (TaKaRa ExTaq DNA polymerase; Takara Bio Inc., Shiga, Japan). The PCR products were purified using a PCR Gel Purification Kit (Qiagen) and a Labo Pass PCR Purification Kit (Cosmo Genetech, Seoul, Korea). PCR products were sequenced by Cosmo Genetech.
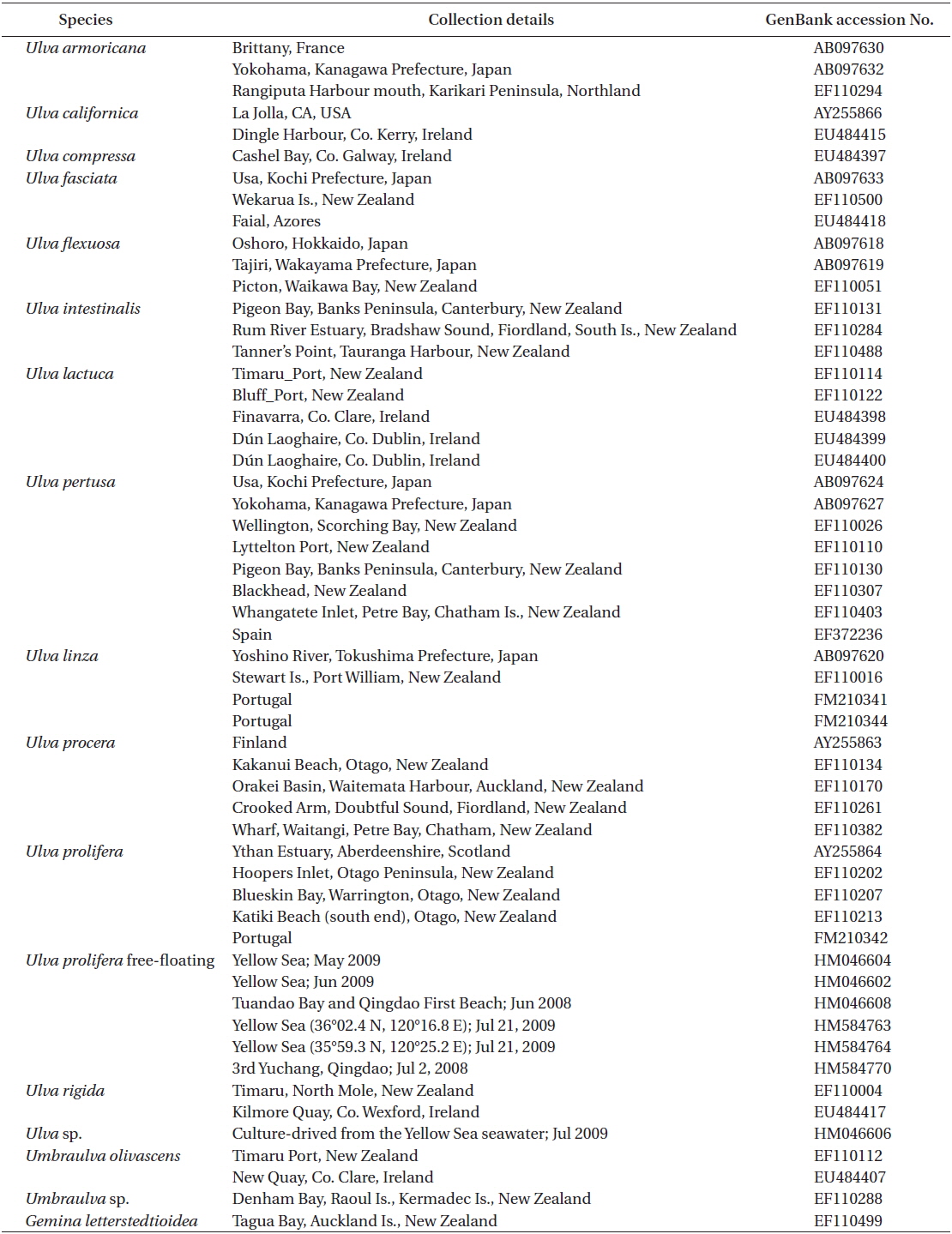
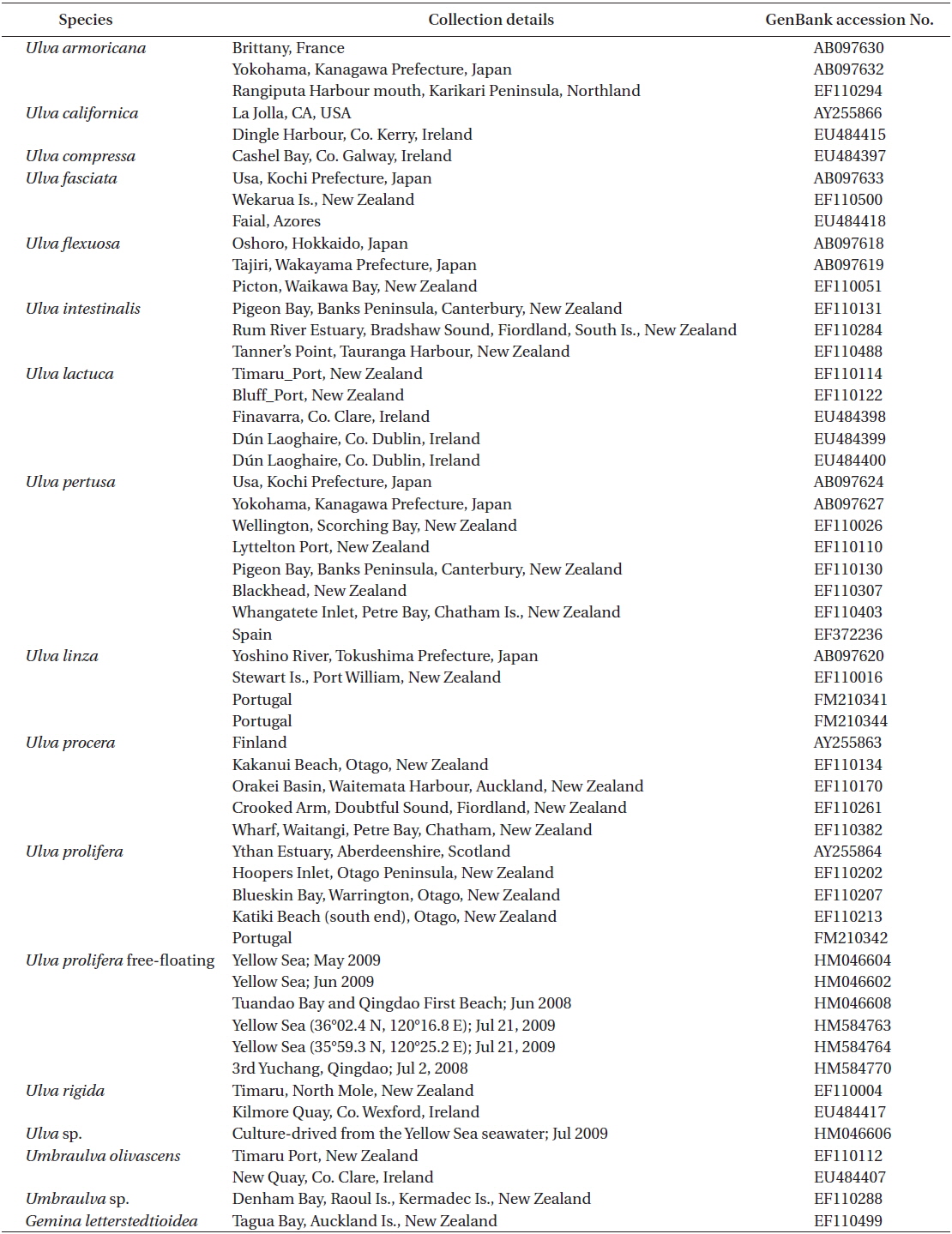
Sequence data were edited using the SeqEd DNA sequence editor software package, and the edited DNA sequence alignment was conducted manually using SeqPup (Gilbert 1995) and MacClade v.4.06 (Maddison and Maddison 2003) after adding other rbcL sequences retrieved from GenBank (Table 3). Bayesian phylogenetic trees were constructed with MrBayes 3.0b4 (Huelsenbeck and Ronquist 2001). The GTR + Г + I model was used and 5,000,000 generations of four chains were run sampling every 100 generations. The neighbor joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) methods were implemented using PAUP 4.0b10 for the Macintosh (Swofford 2002). We used Modeltest 3.7 (Posada and Crandall 1998) for ML and distance analyses to determine a suitable model for our data. The best-fit model was TrN + I + G selected by hLRTs. The ML analysis was completed using the heuristic search algorithm with five random additions. The NJ analysis was subjected to 1,000 rounds of bootstrap resampling, and the MP analysis used 5,000 resampling rounds with 10 random addition replicates per bootstrap replicate.
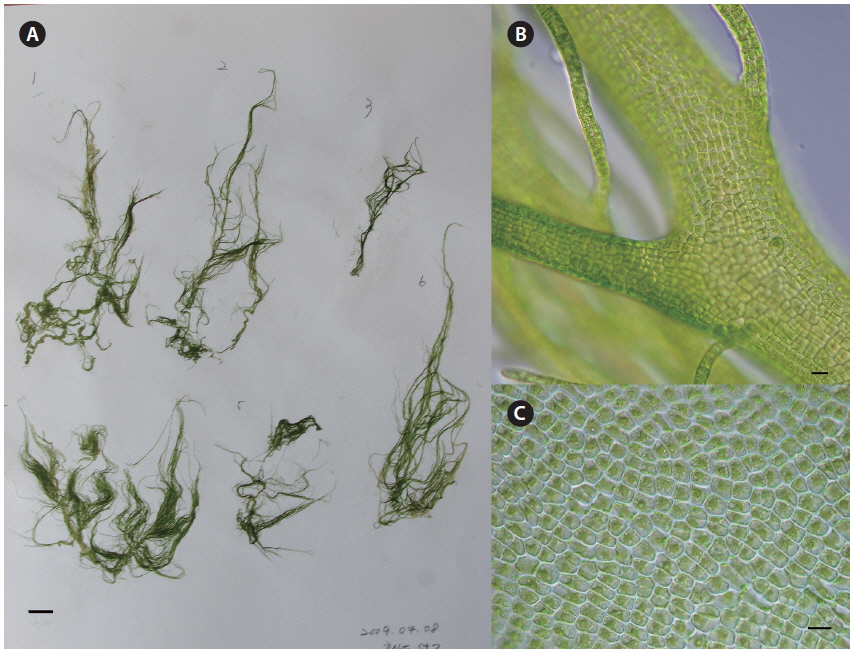
The major bloom-forming species, U. linza, generally showed procera-morphology. This morphotype of U. linza had a narrow tubular shape, was highly branched throughout all thalli, and had central cavities in most thalli and branches (Fig. 1). U. linza thalli were <2 mm wide and had rectangular or quadrangular cells arranged in longitudinal and transverse rows. Each cell had one, or rarely, two pyrenoids.
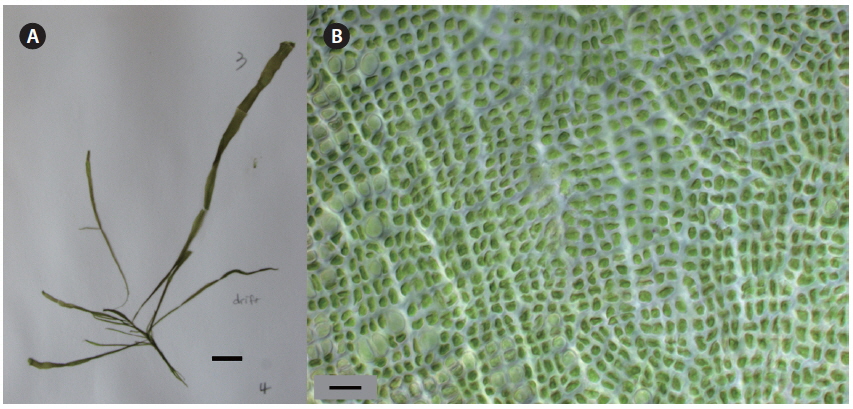
U. compressa had tubular thalli with irregularly spaced regions of greater or lesser inflation (Fig. 2). Some thalli were highly branched or unbranched. The rounded cells were arranged irregularly but were occasionally found in longitudinal rows. The cells had one large pyrenoid.
U. prolifera was very morphologically similar to the procera-morphology of U. linza. U. prolifera thalli were branched intensively with a narrow primary axis and tubular thalli, and all branches had central cavities. Thalli were <2 mm wide, and the quadrangular cells were arranged in longitudinal rows. The cells had one pyrenoid.
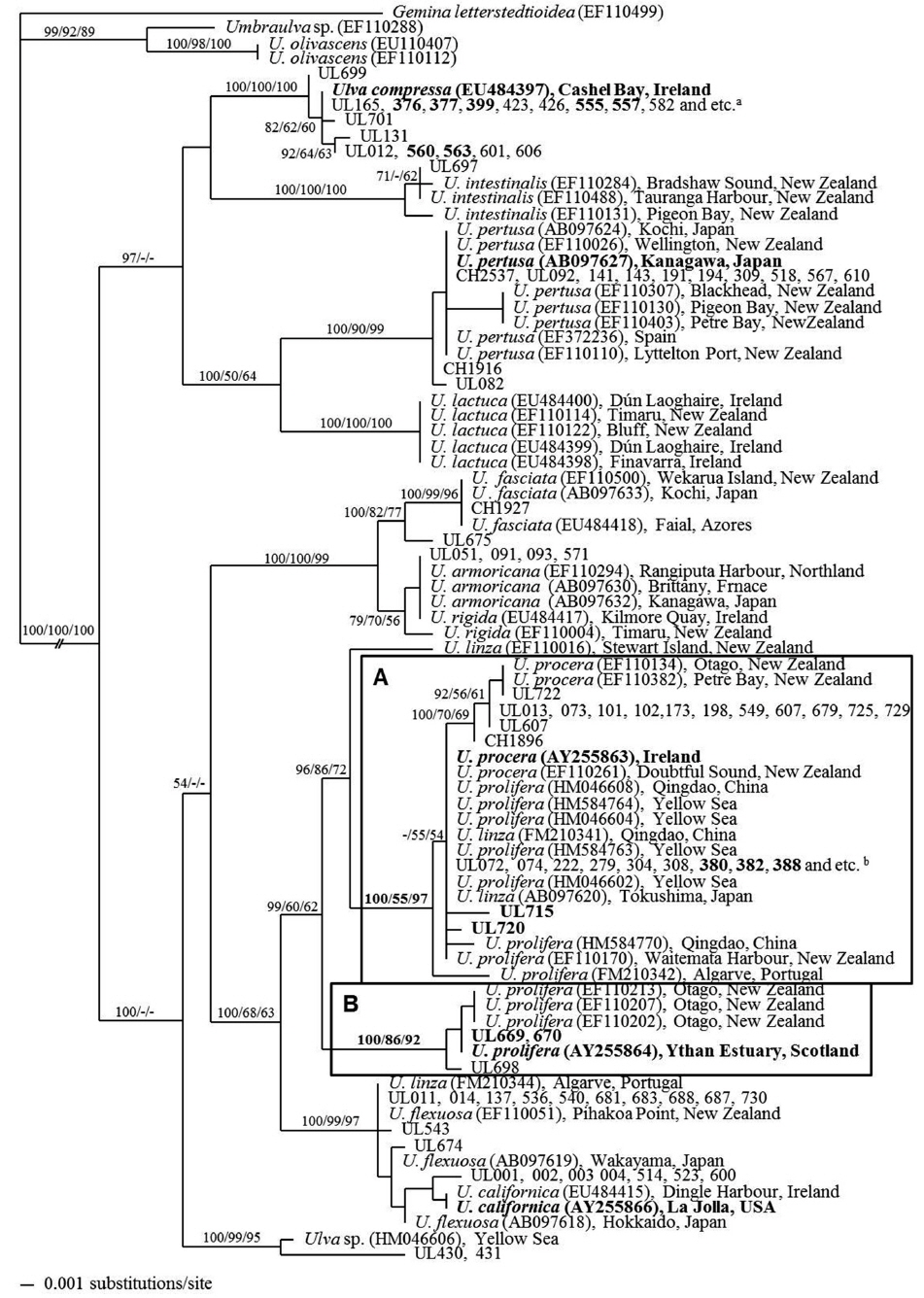
All Ulva spp. belonged to a monophyletic group with strong bootstrap support. A phylogenetic tree based on the rbcL sequence data of Ulva spp. distinguished 10 clades, including one unidentified Ulva sp. collected from the Korean coast (UL430, 431) (Fig. 3). Eight species were identified from samples collected from the Korean and Chinese coasts, and one of them was recorded from the floating green mat off the Qingdao coast (UL669, 670).
[Fig. 3.] Tree constructed with Bayesian inference for the rbcL. Values at branches represent Bayesian posterior probabilities (left), and 1,000 bootstrap replicates for neighbor joining (mid) and 5,000 bootstrap replicates for maximum parsimony (right). Lacking values received less than 50% support. Taxa names in bold type indicate samples collected near type locality of the species. Sample numbers are described in Table 1. Samples in bold type indicate free-floating samples from the Yellow Sea. Bold box A and B indicates Ulva linza group (reported as U. procera and U. prolifera by various authors) and U. prolifera group, respectively. aUL618, 691, 695, 700, CH2538, 2543, 2544. b383, 394, 410, 413, 526, 532, 535, 592, 595, 596, 663, 664, 672, 705, 707.
The floating green mats consisted U. linza, U. compressa, and U. prolifera (Fig. 3). The main floating green mat species in the YS from 2008 to 2011 was U. linza, which was identified by rbcL analysis (e.g., UL380, 382, 388, 715, 720). These samples were identified as U. prolifera in previous studies, which belongs to the same clade as the YS green tide species from 2008 and 2009 (e.g., HM046604, HM584763) (Liu et al. 2010c, Duan et al. 2012). The floating green mats from the Qingdao coast (UL592, 596) and culture-derived samples from the YS (UL663, 664) were identified as U. linza. This clade also included attached Ulva specimens from the Korean and Chinese coasts (e.g., UL072, 308, 549, 607).
A small number of U. compressa specimens were found in the floating green mats from the Korean and Chinese coasts in 2009 (e.g., UL376, 555) (Fig. 3). U. compressa appeared as a distinct monotypic clade but floating and attached specimens were not distinguished in the tree. U. prolifera was also collected from the Qingdao coast floating green mats (UL669, 670) (Fig. 3). The U. prolifera clade, including rbcL data from Scotland (AY255864) and New Zealand (EF110202, EF110207, EF110213), was strongly supported by bootstrap values (Bayesian posterior probabilities, 100%; neighbor joining, 86%; and maximum parsimony, 92%). U. prolifera did not occur on the Korean coasts from 2009 to 2012 and did not form a green tide in the YS.
Based on morphological evidence and the rbcL sequence data of 106 samples collected from the YS, including the east coast of China and southwest coast of Korea, the massive green mats were comprised of three species (U. linza, U. prolifera, and U. compressa). U. linza and U. compressa occurred on both sides of the YS (the east coast of China and the southwest coast of Korea), whereas U. prolifera only occurred along the east coast of China. The green tide-forming species in the YS have been regarded as U. prolifera in many studies (e.g., Wang et al. 2010, Zhang et al. 2011). However, U. linza was the dominant species, and it was mixed with some other Ulva spp. in our study.
The typical form of U. linza is strap-shaped, with oblong, oblanceolate, and sheet-like thalli and ruffled blade margins (Bliding 1963, Koeman 1985, Brodie et al. 2007). However, U. linza does not have a consistent morphology because of extensive phenotypic plasticity induced by the environment (Bliding 1963). Ulva spp. are not always distinguishable based on morphology (Blomster et al. 1998, 1999). We also found that identifying Ulva spp. based on morphological characters, such as thallus form, cell shape, and arrangement, was problematic (Figs 1 & 2). Bloom-forming U. linza in the YS were similar in morphology to that of U. prolifera and U. procera (regarded as a synonym of U. linza) (Tan et al. 1999, Brodie et al. 2007, Heesch et al. 2007). According to Brodie et al. (2007), U. linza has two primary morphological forms, i.e., the linza-morphology and the procera-morphology. Most floating bloom samples from the YS were confirmed to be U. linza the rbcL sequence data, and it manifested the procera-morphology under these conditions (Fig. 1).
The morphological characters of bloom species collected from Korean coastal waters were similar to those of the bloom species from the coasts and offshore of China (Fig. 1). Our the rbcL sequence data showed that not only did the species west and southeast of the YS (e.g., UL380, UL592, UL072, HM046608) occur in the same clade but that both morphological types (i.e., linza- and procera-morphologies) were present within this clade (Fig. 3).
Many studies of the YS green tide species have attempted to clarify the identity of the species and its origin. Early studies that identified the YS green tide species concluded that the bloom species belonged to the LPP complex based on difficulties using internal transcribed spacer (ITS) and rbcL gene sequences (Leliaert et al. 2009, Liu et al. 2010b, 2010c). Although U. procera is included in the LPP complex, U. procera is regarded as a synonym of U. linza; thus, it is more accurate to classify U. procera in Ulva linza-prolifera (LP) complex (Tan et al. 1999, Brodie et al. 2007, Guiry and Guiry 2014). Nevertheless, many researchers concluded that the YS green tide species was U. prolifera based on morphological traits and that it belonged to the LPP complex (e.g., Pang et al. 2010, Wang et al. 2010, Zhang et al. 2011, Liu et al. 2012). However, phylogenetic results show that U. prolifera is in the LP complex, and we believe that many researchers are unaware that the LPP complex is an artificial assemblage.
Wang et al. (2010) concluded that the YS bloom species was U. prolifera based on a ribotype network analysis of ITS sequences in which the bloom-forming material was closely related to Japanese U. prolifera from brackish water, rather than that of Europe and New Zealand. This was despite the fact that this material could be ascribed to the LPP complex based on the ITS gene. Shimada et al. (2008) first suggested the LPP complex originated from an ITS phylogenetic tree because the phylogenetic tree showed no substantive differences between U. linza and U. prolifera. However, the LPP complex in their results did not include U. prolifera samples, and U. prolifera from Europe was separated in another clade from the LPP complex. Therefore, it is possible that Japanese U. prolifera in brackish water was mis-identified.
Hiraoka et al. (2011) noted potential problems identifying the YS bloom species based on the ITS marker. The 5S rDNA spacer sequence within the LPP complex has been used because the 5S rDNA spacer is 10 times more variable than that of the ITS region (Hiraoka et al. 2011, Zhang et al. 2011, Han et al. 2013, Huo et al. 2013). However, these studies did not include specimens from the U. prolifera clade in Europe. Shimada et al. (2008) used the 5S rDNA spacer to elucidate the phylogenetic and phylogeographic relationships of Japanese U. prolifera within the LPP complex from fresh and brackish water. Hiraoka et al. (2011) carried out a hybridization study with Japanese U. linza and two strains of U. prolifera from Qingdao and Japan. U. prolifera from the Qingdao strain hybridized successfully with the Japanese strain, but did not mate with U. linza. They insisted that the YS green tidecausing species was a U. prolifera subspecies based on 5S rDNA spacer sequences and crossing experiments. As mentioned earlier, U. prolifera is identified primarily using morphological traits, and U. prolifera from Japan had the same ITS and 5S rDNA spacer sequences as some samples from Shimada et al. (2008), which may have been based on a mis-identification. Our analysis showed that U. linza and U. prolifera were in distinct clades (Fig. 3). U. prolifera (UL699, 670) in our results belonged to the same clade as U. prolifera samples from Europe (AY255864 and UL698) and New Zealand (EF110202, EF110207, and EF110213) (Fig. 3). Thus, we could not conclude that the major strain causing the green tide was U. prolifera. We hypothesize that the YS green tide species is a new ecotype of U. linza based on reproductive isolation from attached U. linza and their morphological differences.
The algae forming the YS green tide were not unialgal species. This green tide included U. linza as the predominant species. In addition, U. compressa was also found in the floating green mats on the western and southeastern coasts of the YS, even though there was much less material. We also found U. prolifera in the Qingdao coast mats. However, it was difficult to distinguish U. compressa and U. prolifera from U. linza based on morphology, and U. compressa was highly morphologically variable, including tubular, inflated, and branched forms. Floating green mats include several Ulva spp., including U. compressa, U. flexuosa, U. linza, and U. prolifera (Han et al. 2013, Huo et al. 2013). These species may have dispersed from coastal areas and intruded the drifting green mats. Therefore, the floating green mats may include other Ulva spp. after moving offshore.
Massive green algal blooms have occurred annually in the YS since 2007, and the blooms off the southwest coast of Korea subsequently appeared 2-3 months after the massive bloom off the coast of China. The bloom-causing species in Korean coastal waters is the same species as the Chinese U. linza identified by rbcL sequence data, and it has the same procera-morphology. The LPP complex of U. prolifera referred to in previous studies is U. linza, not U. prolifera. Even a type locality U. prolifera sample has not been considered in Ulva phylogenetic studies to date (including this study). Some minor components of the green tide are U. compressa and U. prolifera, and these are mixed with the primary mat forming entity. We conclude that the green tide-forming species is not U. prolifera, but U. linza. According to recent studies from New Zealand, Australia, and the USA, U. prolifera is monophyletic regardless of the genes (Heesch et al. 2009, Kraft et al. 2010, Guidone et al. 2013, Kirkendale et al. 2013). Several Ulva spp. phylogenetic studies regard U. procera as a separate species from U. linza (Heesch et al. 2009, Saunders and Kucera 2010, Kirkendale et al. 2013). However, we conclude that the main bloom-causing species is another ecotype of U. linza with procera-morphology, because U. procera is accepted as a synonym of U. linza.
The YS green algal bloom is an annual environmental problem for China and Korea. There are still unanswered questions regarding the bloom-forming species because of their unusual physiological and ecological characteristics. Given the ecological importance of this bloom, an understanding of the causation and potential management of this nuisance phenomenon must begin with correct identification of the algae involved.