Aquaculture has been practiced in Korea for several hundred years, with a significant contribution from mariculture farming. As such, it is important to maintain a sustainable mariculture industry in order to gain economic and environmental benefits. The mariculture field has encountered significant production losses in recent years due to the occurrence of numerous infectious diseases. Although there is a high demand for rock bream
Follistatin (FST) is a monomeric glycoprotein that is considered an inhibitor of follicular stimulating hormone. FST has the ability to suppress other proteins in a manner similar to that of inhibin (Welt et al., 2002). Moreover, it can antagonize several growth factors, including activins, a group of TGF-β superfamily members (Tortoriello et al., 2001). FST is expressed in all types of tissues and exhibits a wide range of tissue distribution. This diversified tissue expression pattern also provides clues to understanding the primary function of FST, which can effectively modify the paracrine/autocrine roles of activin in a variety of tissues. To date, two major FST subfamilies have been identified (Clabaut et al., 1988). Subfamilies are classified based on the two major characteristics of FST-like proteins: (a) a relatively strong activin binding ability and (b) structural homology to other FST-like proteins. Members of the first subfamily have both properties while members of the second subfamily lack the ability to bind activin.
It is known that FST plays a major role in fish development, since significant levels of FST expression have been detected in a zebrafish model (Bauer et al., 1998). FST was found to be expressed at relatively high levels during advanced stages of embryonic development (Nakamura et al., 1990; Michel et al., 1993; Amthor et al., 2002). Moreover, according to previous studies on activin A, FST is involved in fish reproduction and muscle development. By antagonizing and interacting with the functions of members of the TGF-β superfamily, FST can regulate a variety of TGF-β superfamily proteins in growth- and development-related pathways. Early detection of FST transcripts in sea bream embryos also suggested a possible physiological role in fish development (Funkentein et al., 2009). In addition to activin A, extensive studies on FST suggest its involvement in antagonizing the action of several other members of the TGF-β superfamily. FST itself can also interact with bone morphogenetic proteins (BMPs) (Fainsod et al., 1997; Iemura et al., 1998; Amthor et al., 2002), growth differentiation factor-9 (GDF-9) (Lin et al., 2003), and myostatin [MSTN, or growth differentiation factor-8 (GDF-8)] (Zimmers et al., 2002) in order to abolish their functions during particular growth stages.
Recent studies on FST have provided new insight on its functions, including its involvement in immune-related pathways and developmental physiology. The induction of activin A causes the death of B cells, which are major components of the host defense mechanism (Yu et al., 1987). Additionally, differentiation and proliferation of cells from erythroid lineages was observed following the induction of activin A expression (Broxmeyer et al., 1988). These observations, and its ability to interact with other cytokines such as IL-1 and IL-6 at different regulatory levels, confirm the involvement of activin A in inflammatory pathways (Yu et al., 1987). The interaction of FST with members of the TGF-β superfamily, including activin A, suggests that FST can potentially participate in host immune/inflammatory responses. Moreover, studies under septic conditions (Broxmeyer et al., 1988) showed that because of this FST-activin A interaction, FST must also have a critical role in inflammatory disorders, as it is a stiff inhibitor of activin A. In this study, we have characterized the
>
Identification of the complete RbFST cDNA sequence
The full-length
>
Bioinformatic characterization of RbFST sequences
The open reading frame (ORF) and amino acid sequences of RbFST were derived using DNAssist 2.2. The FST protein sequences from other species were obtained using a BLAST search. These sequences were then used for pair-wise and multiple sequence alignments by utilizing EMBOSS needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/) and ClustalW2, respectively (Thompson et al., 1994). Phylogenetic relationships were determined with Molecular Evolutionary Genetics Analysis (MEGA) software version 5 (Tamura et al., 2011) using the Neighbor-Joining method and bootstrapping values taken from 1000 replicates. Characteristic protein signatures in the RbFST sequence were predicted using the ExPASy-prosite server (http://prosite.expasy.org) and the NCBI conserved domain database (CDD) (Marchler-Bauer et al., 2011). Some physicochemical properties of RbFST were determined using the ExPASy ProtParam tool (http://web.expasy.org/protparam).
>
Experimental animals and tissue collection
To examine the immune response of
>
Total RNA extraction and cDNA synthesis
Total RNA was extracted from rock bream tissues that had been pooled from three fish, from both the control and challenged groups, using Tri ReagentTM (Sigma). After quantification by UV spectrophotometry (optical density at 260 nm), the total RNA samples were diluted to 1 μg/μL and used to perform cDNA synthesis using the PrimeScriptTM cDNA Synthesis Kit (TaKaRa Bio, Japan), according to the manufacturer’s instructions. Finally, newly synthesized cDNA was diluted 40-fold (total volume: 800 μL) and stored at -20°C until use.
>
RbFST mRNA expression analysis by quantitative real-time PCR (qPCR)
qPCR was performed using the DiceTM TP800 Real-Time Thermal Cycler System (TaKaRa) in a 15-μL reaction volume containing 4 μL of diluted cDNA, 7.5 μL of 2 × TaKaRa
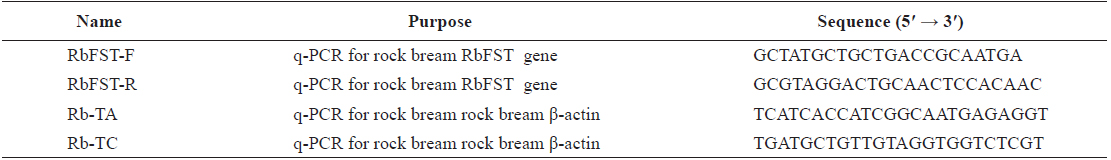
[Table 1.] Oligomers used in this study
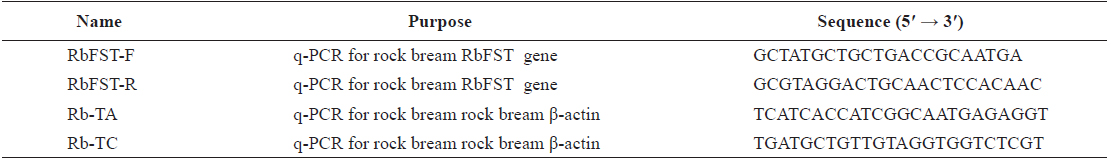
Oligomers used in this study
>
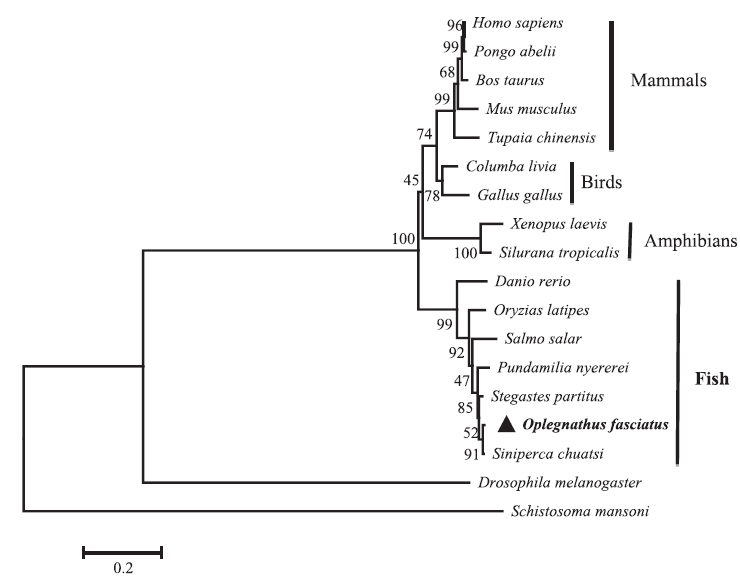
Sequence profile and phylogenetic relationship of RbFST
The
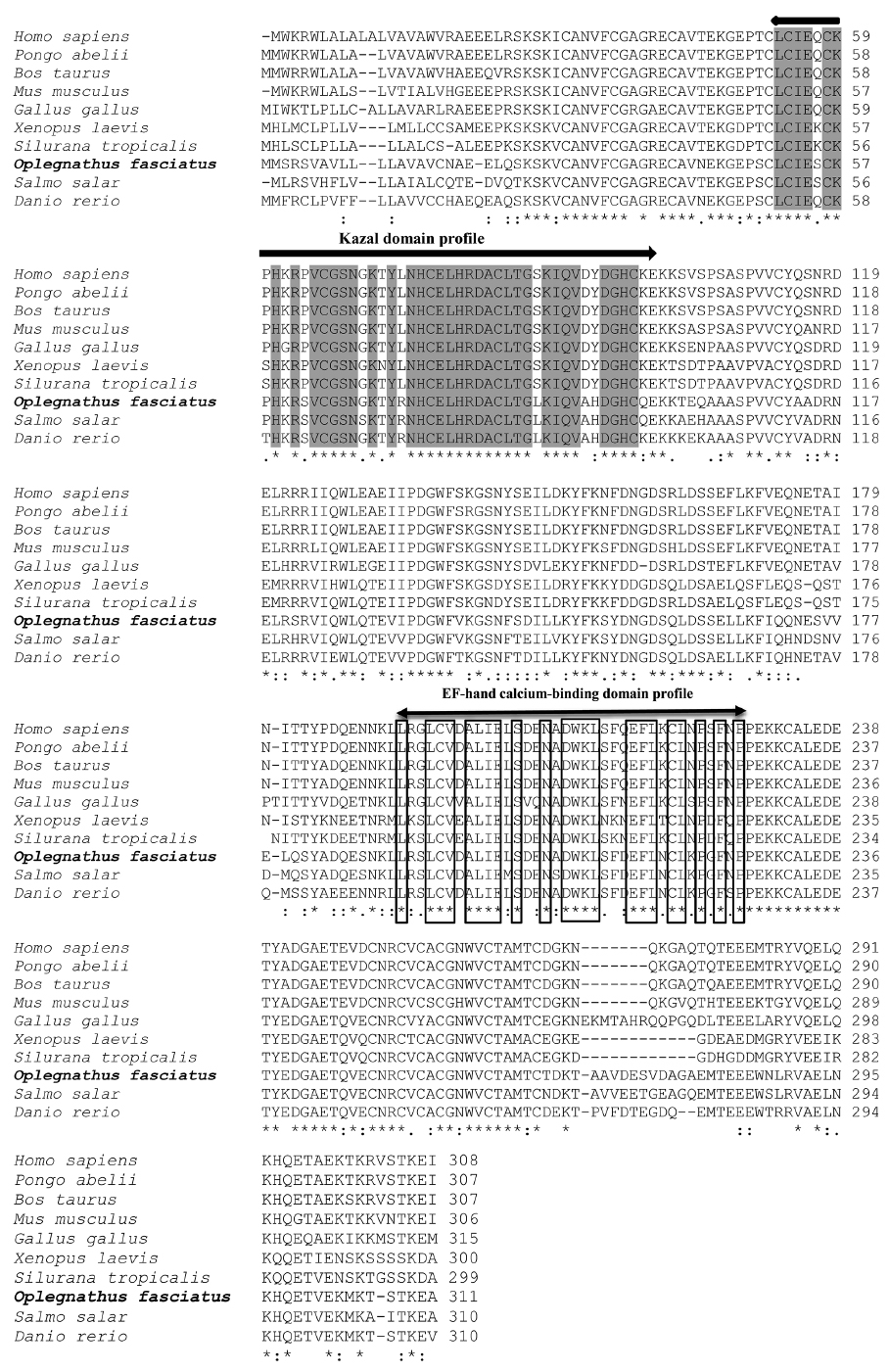
Pair-wise and multiple sequence alignments revealed that the putative RbFST protein shares characteristic features with FST protein orthologs from other species. Multiple sequence alignment (Fig. 2) showed the presence of a highly conserved Kazal domain, which is a feature of FSTs. The EF-hand, calcium binding domain is also identified in FSTs as a conserved domain among several species, including rock bream. Pairwise sequence comparisons showed that
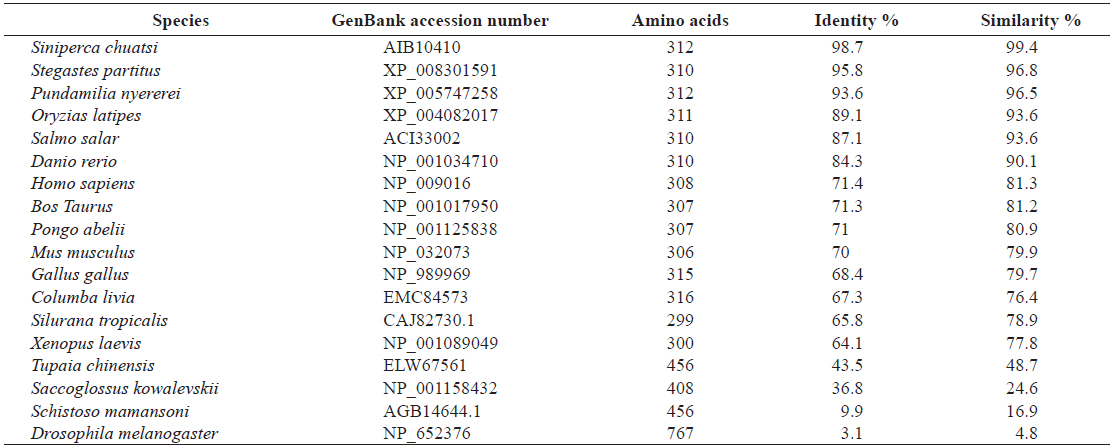
[Table 2.] Amino acid identity and similarity of RbFST entire sequence to other homologues
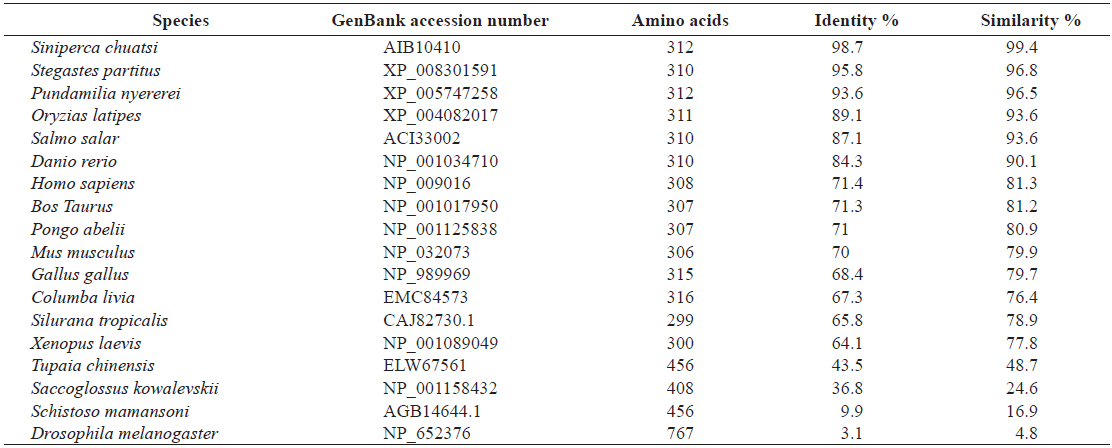
Amino acid identity and similarity of RbFST entire sequence to other homologues
>
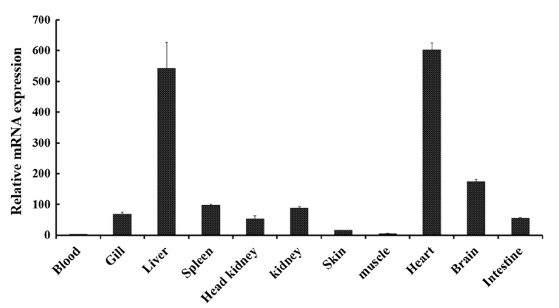
Tissue-specific mRNA expression profile of RbFST
In order to determine the transcriptional profile of
The main role of FST in heart tissues remains unclear and studies are being conducted to identify the involvement of FST in cardiac conditions (Ogura et al., 2012; Miyabe et al., 2014). Even though the expression of
Prominent expression levels of
>
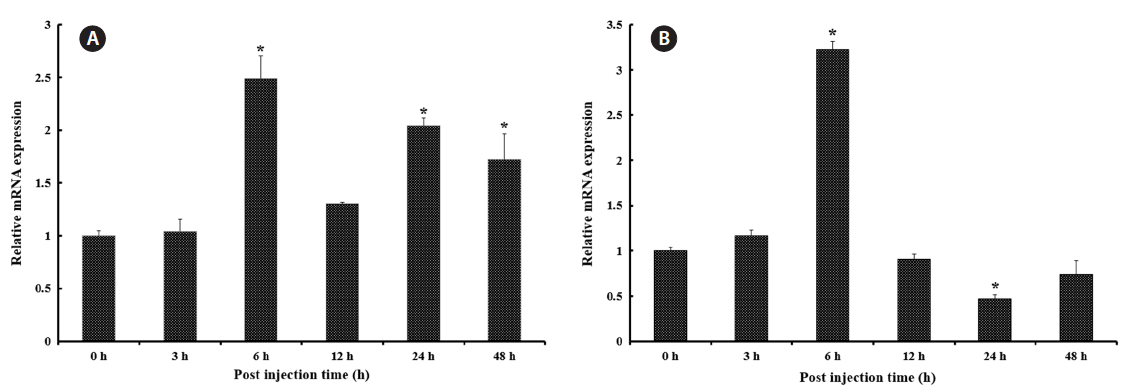
Transcriptional modulation of RbFST upon pathogen injection
In both vertebrates and invertebrates,
As shown in Fig. 5A, after healthy rock breams were challenged with
We have characterized a member of the FST-family of proteins from rock bream (RbFST) at the molecular level and determined its expression levels in different tissues under physiological conditions, revealing ubiquitous expression. Moreover,