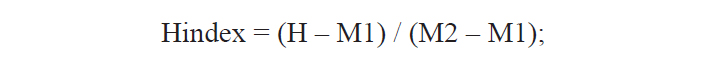As in plants, hybridization and introgression play important roles in the evolution of fish (Hubbs, 1955; Dowling and DeMarais, 1993; Scribner et al., 2001). They represent the driving force behind the evolution of new lineages with the potential to rapidly adapt to new habitats (DeMarais et al., 1992; Gerber et al., 2001; Grant and Grant, 1992). However, hybridization can also lead to extinction of rare indigenous species and presents challenges for the formulation of a biological species concept and definition, the reconstruction of phylogenetic trees and the practice for biological conservation (Grant and Grant, 1992; Rhymer and Simberloff, 1996; Allendorf et al., 2001). Natural hybridization occurs across diverse piscine taxa (Verspoor and Hammar, 1991) and is quite common in cyprinid fishes (Hubbs, 1955; Gerber et al., 2001; Scribner et al., 2001; Ünver et al., 2008). Hybridization between fish species frequently occurs in nature resulting from the intrinsic properties of the species, external fertilization and weakly developed reproductive isolating mechanisms, as well as natural or man-made causes, such as disparity in the number of parent species, habitat disturbance and fragmentation, the introduction of non-native species, and climate change (Hubbs, 1955; Rhymer and Simberloff, 1996; Allendorf et al., 2001; Scribner et al., 2001; Šorić, 2004; Ünver et al., 2008; Kwan et al., 2014).
Morphological analysis is a conventional approach to identification of naturally occurring hybrids because of the intermediate phenotypes. However, morphology alone is not always sufficient to determine the hybrid status and to trace the parent species (Avise and Saunders, 1984; Allendorf et al., 2001; Scribner et al., 2001). Analyses of genetic markers with different evolutionary histories provide convincing evidence for hybrid status and can help determine the extent and direction of hybridization (e.g., Avise and Saunders, 1984; Dowling et al., 1989; Avise et al., 1990; Albert et al., 2006; Chelomina et al., 2008). In practice, the combined application of nuclear DNA (nDNA) and mitochondrial DNA (mtDNA) sequences has proven useful in determination of hybridization in fish species (e.g., Sonnenberg et al., 2007; Lee et al., 2009; Yoon et al., 2009). These techniques are simple and easy to implement as the result of recent advances in molecular techniques, such as polymerase chain reaction (PCR), DNA sequencing, etc.
The mitochondrial genome is haploid, located in the cytoplasm and is maternally inherited without recombination. Given these distinct properties, analyses of mtDNA sequences provide an opportunity to determine the direction of gene flow in a hybrid cross (Avise and Saunders, 1984; Moritz et al., 1987; DeMarais et al., 1992; Rhymer and Simberloff, 1996). However, the information provided by mtDNA alone is limited due to its strict maternal mode of inheritance. Thus, to determine the directionality of species hybridization, mtDNA analysis is often used in conjunction with analysis of a nuclear gene, which is inherited biparentally or in a simple Mendelian fashion. Hybrid state is reliably identifiable in monomorphic (homozygous) versus polymorphic (heterozygous) states; i.e., each pure parent species possesses a clear DNA sequence with monomorphic base positions, whereas their hybrids exhibit polymorphisms different to the parent (Scribner et al., 2001; Sonnenberg et al., 2007).
The striped shiner Pungtungia herzi Herzenstein and the stone morocco (or the false dace) Pseudorasbora parva Temminck and Schlegel (Cypriniformes: Cyprinidae) are widely distributed in East Asia and coexist in the river basins of Korea (Kim and Park, 2002; Kim et al., 2005). A natural hybrid of a probable mating between Pu. herzi and Ps. parva was captured during a survey of ichthyofauna in Korea. Morphological characters as well as nDNA and mtDNA sequences were analyzed to verify the hybrid state and determine the parentage.
A single natural hybrid (n = 1; standard length: 64.7 mm) between Pu. herzi and Ps. parva was captured with a spoon net (mesh size: 4 × 4 mm) on 21 April 2010 in the Geumho River, a tributary of the Nakdong River basin in Korea (Fig. 1). Specimens of its supposed parent species, Pu. herzi (n = 16) and Ps. parva (n = 11) were also sampled from the same tributary. The specimens were deposited in the freshwater fish collection of the Department of Marine Biotechnology of Soonchunhyang University (SUC; Asan, Korea). Detailed sampling information is provided in Table 1.
A total of 25 morphological characters were used in the calculation of the hybrid index (Hindex) to measure the degree of phenotypic intermediacy of the hybrid in comparison to its parent species. The number of fin rays and vertebrae were counted using a soft X-ray photographer (Hitex HA-80; Hitex, Osaka, Japan). Total of six meristic and 19 morphometric characters were counted or measured as described by Hubbs and Lagler (1964) (Table 2). The mean of Pu. herzi was set to zero, and the mean of Ps. parva to 100 to calculate the Hindex, which was computed using the following formula (Nikoljukin, 1972):
where H is the numerical value of a character of the hybrid, and M1 and M2 are those of the same character in Pu. herzi and Ps. parva, respectively.
A section of the pectoral or anal fin was excised from each specimen to extract genomic DNA (gDNA). The tissue was incubated with TNES-Urea buffer (10 mM Tris-HCl, pH 8.0; 125 mM NaCl; 10 mM EDTA, pH 8.0; 1% SDS; 8 M urea) (Asahida et al., 1996) containing 100-μg proteinase K (Sigma-Aldrich, St. Louis, MO, USA) at 37℃ for one day, followed by separation of the aqueous phase with a phenol : chloroform : isoamyl alcohol (25 : 24 : 1) solution, and ethanol precipitation according to Sambrook and Russell (2001). The extracted gDNA was resuspended in 1× TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0). Its quantity and quality were checked using a spectrophotometer, NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE, USA) and by electrophoresis in a 0.7% agarose gel after staining with GelRed™ Nucleic Acid Gel Stain (Biotium, Hayward, CA, USA).
To amplify a complete fragment of the mitochondrial cytochrome b gene (mt-cyb) and a partial fragment of the nuclear recombination activating protein gene 1 gene (rag1), a PCR was carried out in a 20-μL reaction volume using AccuPower® PCR Premix (Bioneer, Daejeon, Korea), including 50-ng gDNA and 0.2 μM forward and reverse primers [trnE-14317f (5′-GAYTTGAAGAACCAYCGTTG-3′) and trnP-15601r (5′-ATTHTGGCTTTGGGAGYCAG-3′) for the mt-cyb, newly designed in this study, and RAG1-1495f3 (5′-CAGTAYCAYAAGATGTACCG-3′) and RAG1-3067r (5′-TTGTGAGCYTCCATRAACTT-3′) for the rag1 (Kim and Bang, 2010)]. The PCR was conducted using the following thermal cycling profile in a DNA Engine DYAD™ Peltier Thermal Cycler (MJ Research, Waltham, MA, USA): an initial denaturation at 94℃ for 3 min, 35 cycles of denaturation at 94℃ for 30 s, annealing at 55℃ for 30 s and elongation at 72℃ for 1 min. The reaction was completed with a final elongation at 72℃ for 7 min. The PCR product was purified with the AccuPrep® PCR Purification Kit (Bioneer).
The purified PCR product of the rag1 of the hybrid fish was cloned into the pGEM®-T Easy Vector (Promega, Madison, WI, USA) and transformed into competent cells (Escherichia coli XL1-Blue MRF’; Stratagene, La Jolla, CA, USA) according to Chung et al. (1989). A total of 12 white E. coli colonies were randomly picked, and their plasmid DNAs were extracted using the alkaline lysis method (Sambrook and Russell, 2001).
After cycle sequencing using the ABI PRISM® BigDye™ Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA), the purified PCR products and plasmid DNAs were sequenced by Macrogen (Seoul, Korea), a commercial company, on an ABI 3730xl DNA Analyzer (Applied Biosystems) using both sets of PCR primers. The sequences analyzed in this study were deposited in GenBank under accession numbers KP053615–KP053620.
DNA sequences of the rag1 and mt-cyb of the natural hybrid and its parents (Pu. herzi and Ps. parva) analyzed in this study, and those of closely related gobionine species retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) were aligned with ClustalW in BioEdit 7.0.5 (Hall, 1999). Sarcocheilichthys variegatus microoculus (GenBank accession numbers EU711157 and AB054124, respectively) and Sarcocheilichthys parvus (EU409625 and EF193422, respectively) were used as outgroups to reconstruct the phylogenetic trees. The nucleotide sequence matrices for the rag1 and mt-cyb consisted of 1,488 and 1,140 base pair (bp), respectively.
Phylogenetic analyses were carried out in PAUP* 4.0b10 (Swofford, 2002). A neighbour-joining (NJ) tree was reconstructed with the Kimura 2-parameter model, and a maximum-parsimony (MP) tree with the heuristic search option with the random addition of sequences (10 replicates) and tree-bisection-reconnection (TBR) branch swapping. The robustness of the tree topologies was evaluated by bootstrap analyses for NJ and MP analyses with 1,000 pseudoreplicates.
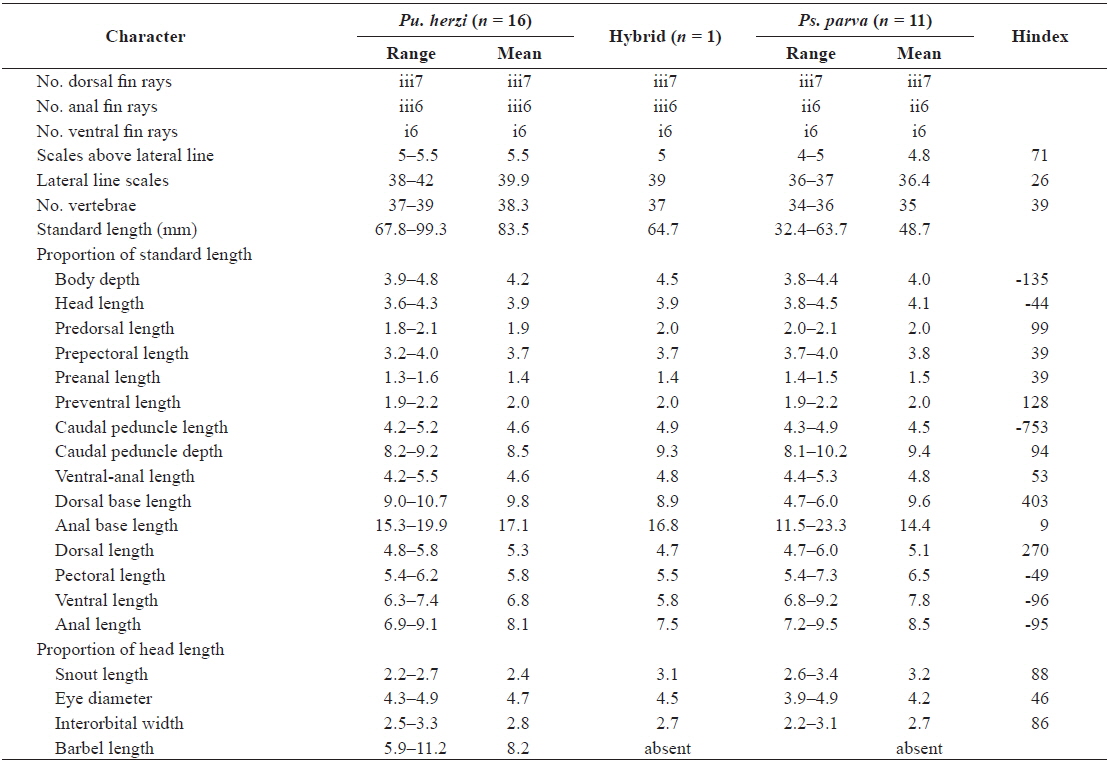
The morphological data for six meristic and 19 morphometric characters along with their Hindex values are presented in Table 2. A Hindex of 50 indicates an exact phenotypic intermediacy between the two parent species. The hybrid was considered to resemble Pu. herzi when the Hindex was < 30, and Ps. parva when the Hindex was > 70; it was considered to be intermediate between the two species when the Hindex was between 30 and 70 (Šorić, 2004).
Of the meristic characters, the number of dorsal and ventral fin rays did not differ among the hybrid and parent species, but the number of anal fin rays of the hybrid and Pu. herzi (iii6) were different than those of Ps. parva (ii6). The scales above the lateral line of the hybrid were more similar to Ps. parva (Hindex = 71), whereas the lateral line scales were more similar to Pu. herzi (Hindex = 26), and the number of vertebrae was intermediate (Hindex = 39). Of the morphometric characters, the anal base length of the hybrid was closer to Pu. herzi (Hindex = 9), while the predorsal length, caudal peduncle depth, snout length and interorbital width were closer to Ps. parva (Hindex = 86–99). Four characters, including the prepectoral length, preanal length, ventral-anal length and eye diameter of the hybrid, were intermediate (Hindex = 39–53). The remaining nine morphometric characters deviated from both parent species.
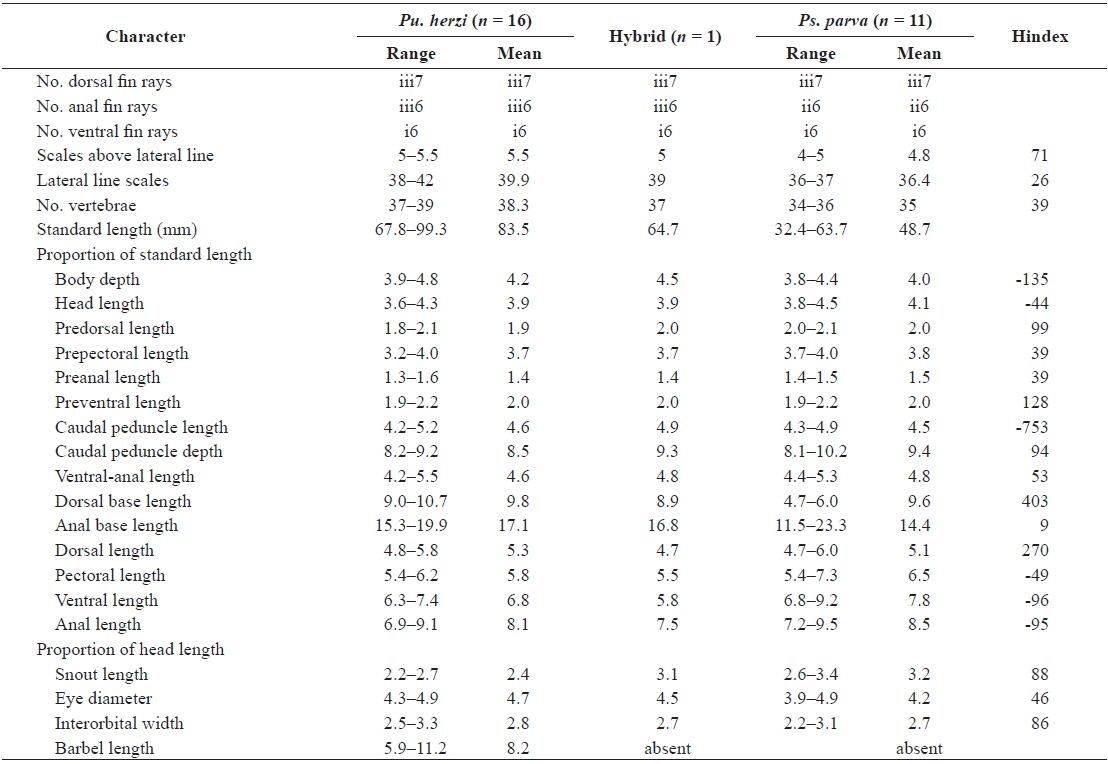
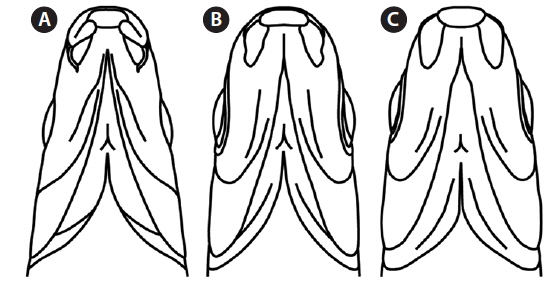
Mouth shape is an important morphological character that distinguishes two parent species (Mori, 1935). Pungtungia herzi has a thick, horseshoe-shaped mouth with a pair of barbels and its upper lip is longer than the lower lip, while Ps. parva has a short, thin upper lip in comparison to the lower lip and lacks barbels (Fig. 2). The natural hybrid possessed thin and identical upper and lower lips lacking barbels.
Partial DNA sequences of the single-copy nuclear rag1 of the natural hybrid and its supposed parent species were analyzed to verify the hybrid state. Both parent species were monomorphic; the electropherograms consisted of unique single peaks at all 1,488-bp positions with 41-bp differences (Fig. 3), showing 97.2% overall identity. Meanwhile, the natural hybrid was polymorphic; its electropherogram displayed double peaks at all 41-bp positions, along with one additional peak at position 711. The hybrid showed an identical ratio of double peaks at the polymorphic base positions where the parent species differed (Fig. 1).
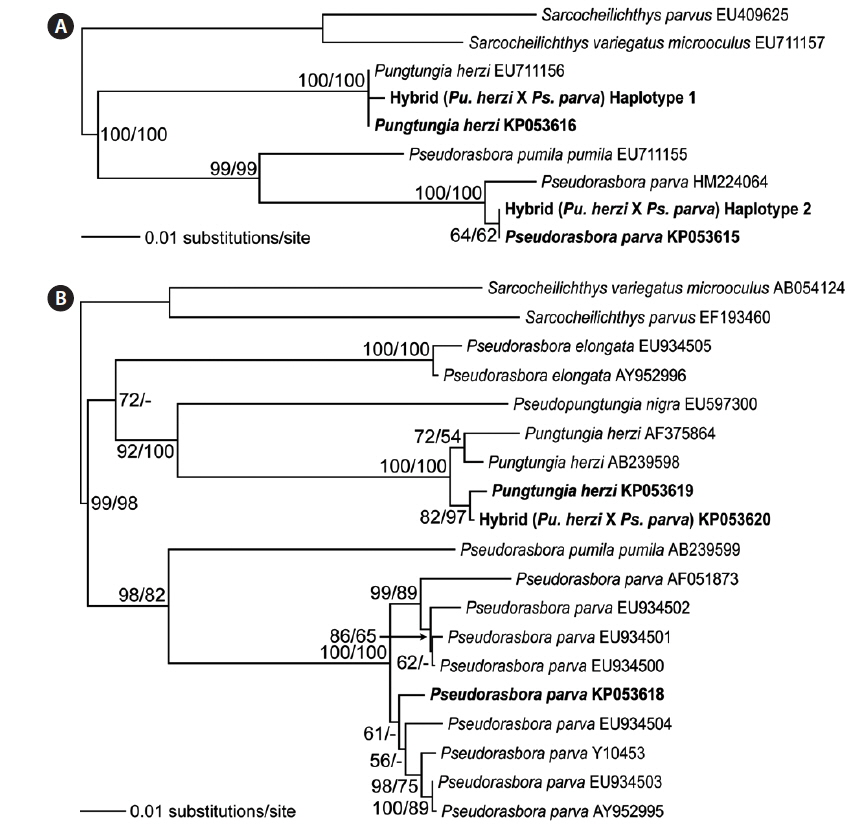
Twelve randomly chosen PCR clones of the rag1 were sequenced to confirm the hybrid status at the polymorphic base positions. The results confirmed the presence of two distinct haplotypes. Seven of the PCR clones corresponded to Pu. herzi, and five to Ps. parva. The consensus sequences of the haplotypes showed 99.9–100% and 99.1–100% identities to those of the parent species, Pu. herzi and Ps. parva, respectively. In the phylogenetic tree, each haplotype consistently clustered with one of the parents, with the highest bootstrap supports (Fig. 4A). Thus, the hybrid possessed two heterogeneous copies of the rag1, one from each parent species.
Analyses of the complete nucleotide sequences of the mitochondrial mt-cyb of the natural hybrid and its supposed parents revealed the direction of hybridization. Pungtungia herzi and Ps. parva displayed 162–180-bp differences out of 1,140-bp positions, showing 83.7–85.7% identities. There were only 5–22-bp differences (97.6–99.5% identities) between the hybrid and P. herzi, whereas there were as many as 158–168-bp differences (85.2–86.1% identities) between the hybrid and Ps. parva. In the phylogenetic tree, the hybrid consistently clustered with Pu. herzi with the highest bootstrap support, showing the closest affiliation with the specimen collected in this study from the Nakdong River basin with 82% and 97% bootstrap supports in the NJ and MP trees, respectively (Fig. 4B). However, it showed a clear phylogenetic distinction from Ps. parva.
A natural hybrid of a probable intergeneric mating between Pu. herzi and Ps. parva was captured in the Geumho River, a tributary of the Nakdong River basin in Korea, and its hybrid state and parentage were verified, based on morphological characters and genetic markers. The putative hybrid displayed intermediate traits in some morphometric and meristic characters, although polarity was only observed in one parent or the other, and vigor from the parent species was also evident.
The nuclear rag1 indicated mixed inheritance in the hybrid with respect to its two parent species. Specifically, the hybrid had polymorphic base positions in its DNA sequence, one of which was inherited from Pu. herzi and the other from Ps. parva, in contrast to the base monomorphic positions in the parent species. In addition, the mitochondrial mt-cyb of the hybrid showed the highest sequence identity to Pu. herzi, implying that this was the maternal species. Thus, the genetic markers strongly suggested the maternity of Pu. herzi and paternity of Ps. parva for the natural hybrid. In addition, the hybrid was considered a F1 hybrid, based on the identical ratio of double peaks at the polymorphic base positions, at which the parent species differed. The single-copy rag1 used in this study (Kim and Bang, 2010) was apparently advantageous over a multi-copy ribosomal RNA gene (Sonnenberg et al., 2007) for determining hybrid status, because the former is inherited in a simple Mendelian fashion.
Pungtungia herzi and Ps. parva are sympatric, but have different habitat preferences and spawning grounds. The former lives on stones or rocks and spawns in host nests (brood parasitism) or crevices between stones and rocks (Yamane et al., 2009), whereas the later inhabits aquatic vegetation and spawns on various substrata such as plants, stones and mollusk shells (Gozlan et al., 2010; Witkowski, 2011). In addition, Ps. parva exhibits aggressive paternal courtship (Konishi and Takata, 2004). These ecological differences and behavioural characteristics appear to contribute to a pre-mating reproductive barrier. However, the spawning periods of Pu. herzi and Ps. parva overlap from April to May in Korea (Kim and Park, 2002; Kim et al., 2005), which may enable mating. In addition, they show close taxonomic (Banarescu and Nalbant, 1973; Kim, 1984; Kim and Kang, 1989) and molecular phylogenetic (Yang et al., 2006; Liu et al., 2010) relationships among species of the Gobioninae, and have the same number of chromosomes (Ojima et al., 1972). Furthermore, Pu. herzi was reported to engage in intergeneric mating with Pseudopungtungia nigra (Kim et al., 1991), and Ps. parva with congeneric Pseudorasbora pumila pumila (Koga and Goto, 2005). Taken together, these properties of the two parent species may increase the probability of intergeneric hybridization as presented in this study. Unfortunately, no other hybrid specimens were found in the region where the hybrid in this study was captured (S.J. Cho, personal observation). Thus, at present, the occurrence of natural hybrids between Pu. herzi and Ps. parva appears to be a rare phenomenon resulting from the accidental contact of gametes.