Starry flounder,
The Na+-K+-2Cl− cotransporter (NKCC) is a member of the cation-chloride cotransporter family that mediates electroneutral movement of Na+ and K+ and is tightly coupled to movement of Cl− across cell membranes. NKCC occurs as a secretory isoform (NKCC1) and an absorptive isoform (NKCC2) (Payne and Forbush, 1994; Xu et al., 1994). In mammals, NKCC2 is expressed mainly in the apical membrane of epithelial cells in the thick ascending loop of Henle where it acts to reabsorb a large amount of NaCl to dilute urine forming in the tubule lumen (Nielsen et al., 1998; Markadieu and Delpire, 2014). NKCC2 dysfunction exerts negative consequences in mammals. In humans, inactivating mutations in the gene coding NKCC2 cause Bartter syndrome type I, which features severe volume depletion, hypokalemia, metabolic alkalosis, and hypercalciuria (Markadieu and Delpire, 2014). Mammalian NKCC2 is the main pharmacological target of loop diuretic drugs used to treat edema (Markadieu and Delpire, 2014).
Piscine NKCC has been suggested to be involved in intestinal salt absorption in several fish species, including European eel,
Starry flounder are a candidate aquaculture species to substitute for olive flounder,
Despite its non-hematopoietic derivation, the intestinal tract functions as a critical defense barrier to the external environment, in addition to being the site of nutrient digestion and osmoregulation (Pastorelli et al., 2013). Ion transport disturbances in mammals have been associated with cellular dysfunction, intra and extracellular edema, and abnormalities in epithelial surface liquid volume (Eisenhut, 2006). However, the functions of teleost NKCC2 genes during immune or bacterial challenge have not been investigated.
Despite the importance of NKCC2 in intestinal function, the molecular mechanism by which this cotransporter is regulated in health and disease is poorly understood. The objective of this study was to characterize the genetic determinants of the NKCC2 isoform from starry flounder and to scrutinize its expression pattern in response to a
Apparently healthy starry flounder (weight, 12.4 g) were obtained from a local Korean fish farm. Fish were reared in 600-L tanks with flow-through seawater maintained at 20℃. The fish were fed a commercial diet once daily before the onset of the experiment. Fish were anesthetized with 0.2% 2-phenoxyethanol for sampling, and body weights were measured. The intestine was split into the anterior intestine, posterior intestine (also referred to as the middle intestine), and rectum based on external appearance (Kim et al., 2008).
>
Molecular cloning of starry flounder NKCC2
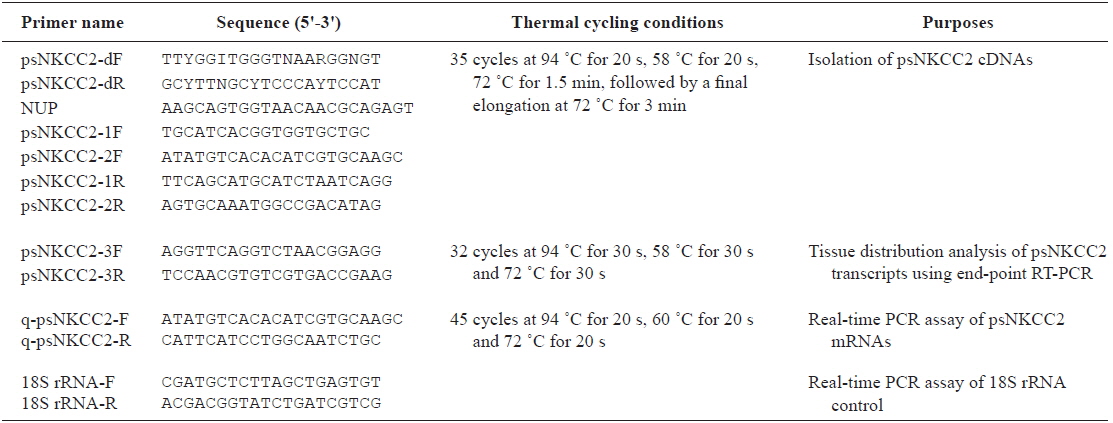
Total RNA was extracted from the intestinal segments using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After a 15-min DNase I (Invitrogen) treatment, total RNA (1 μg) was reverse transcribed using the SMARTTM RACE cDNA amplification kit (Clontech, Palo Alto, CA, USA) according to the manufacturer’s instructions. The degenerate primers to obtain the cDNA fragment encoding NKCC2 were designed based on available information in other vertebrate species. Reverse transcription-polymerase chain reaction (RT-PCR) analysis was carried out using a degenerate primer set (see Table 1 for primers). The resulting PCR products were electrophoresed on a 2% agarose gel and subcloned into the pGEM®-T vector (Promega, Woods, WI, USA) for the sequencing analysis. After determining the partial cDNA sequences, the SMARTTM RACE cDNA amplification kit (Clontech) was used to isolate the full-length cDNA according to the manufacturer’s instructions. The 5’-rapid amplification of cDNA ends (RACE) and 3’-RACE products were produced in primary/nested PCR reactions using specific psNKCC2-1F/2F and psNKCC2-1R/2R primer sets for the NKCC2 gene with an adaptor primer in the kit. RACE products were TA-cloned and sequenced as described above. The nucleotide sequences were determined by analyzing more than five clones for the NKCC2 gene to obtain a representative cDNA sequence.
[Table 1.] Oligonucleotide primers used in this study
Oligonucleotide primers used in this study
>
Sequencing and phylogenic analyses
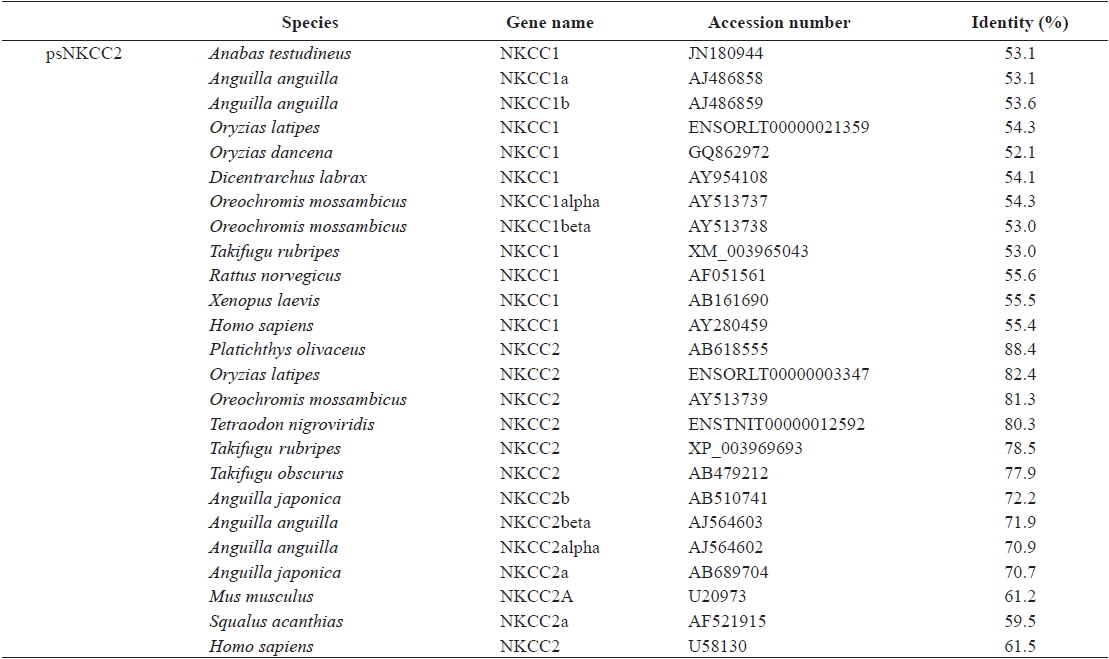
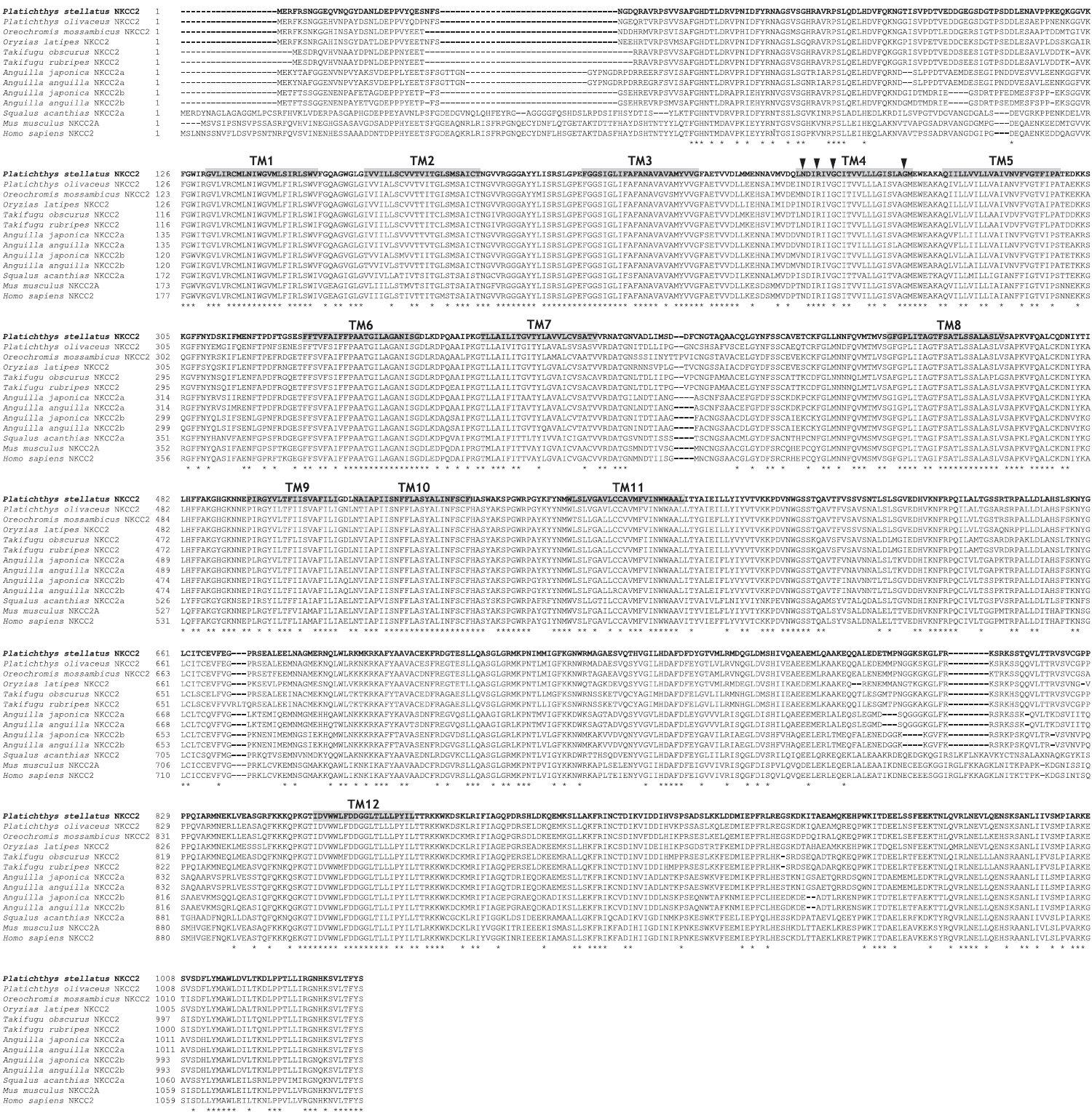
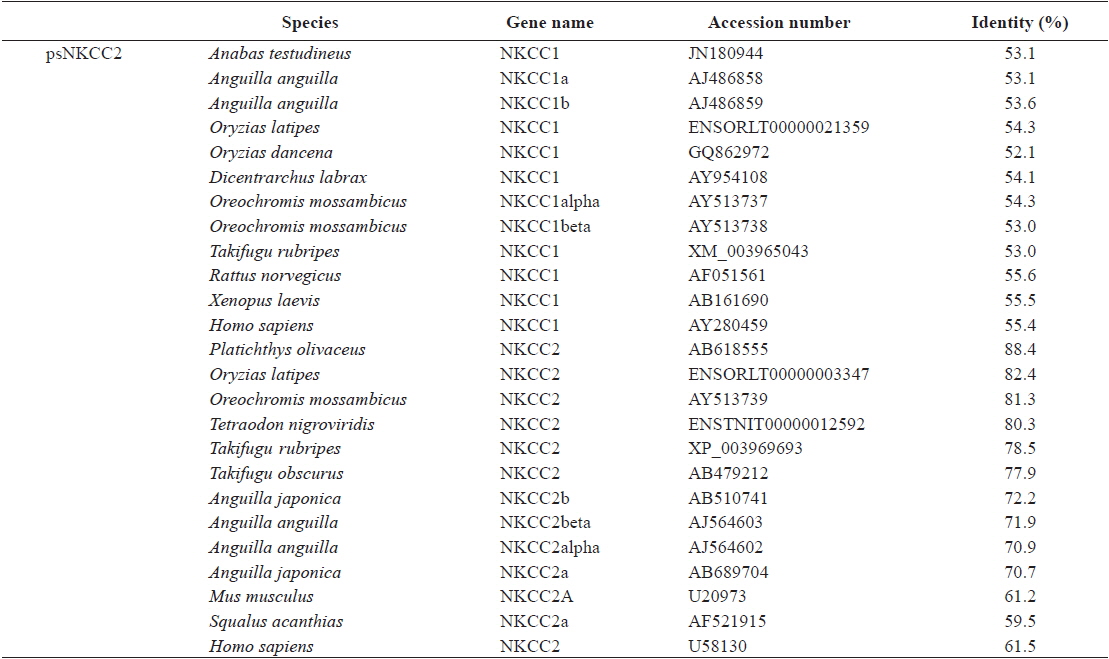
The open reading frame (ORF) analysis of the NKCC2 cDNA sequence was performed using the NCBI web service ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). The molecular weight and theoretical isoelectric point (pI) values of the deduced amino acid sequence were analyzed using the ExPASy ProtParam tool (http://web.expasy.org/protparam/). To determine the degree of homology of the starry flounder NKCC2 isoform with other vertebrate orthologs, we retrieved the NKCC2 gene cDNA sequences of other species from BLAST and/or Ensembl genome database (http://asia.ensembl.org/index.html) searches. Multiple sequence alignments were generated by CLUSTAL W (Thompson et al., 1994). The phylogenic tree was constructed using Molecular Evolutionary Genetics Analysis (MEGA) program (ver. 5.2) with the neighbor-joining method. GenBank accession numbers or Ensembl codes for the NKCC1/NKCC2 sequences employed for the multiple sequence alignments and the phylogenic tree are shown in Table 2. Reliability of the internal tree branches was assessed by 1,000 bootstrap replications. The topology of the NKCC2 deduced amino acid sequences was predicted using the TMHMMfix software (http://www.sbc.su.se/~melen/TMHMMfix/).
[Table 2.] Protein sequence identities of starry flounder NKCC2 with other orthologs
Protein sequence identities of starry flounder NKCC2 with other orthologs
>
Tissue distribution assay of the NKCC2 transcripts
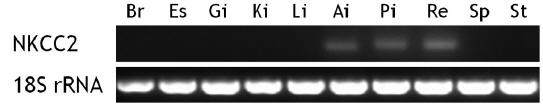
The brain, esophagus, gill, intestine (anterior and posterior intestines and rectum), head kidney, liver, spleen, and stomach were removed from starry flounder under non-stimulated conditions, and total RNA was extracted from the tissues as described above. After a 15-min DNase I treatment, an aliquot of total RNA (2 μg) was reverse-transcribed using the Superscript II First-strand Synthesis System for RT-PCR (Invitrogen). End-point RT-PCR was performed to determine basal expression of the NKCC2 transcripts. The primer pairs for NKCC2 and 18S rRNA were psNKCC2-3F/3R (amplicon = 152 bp) and 18S rRNA-F/R (amplicon = 313 bp), respectively.
>
Experimental Streptococcus parauberis challenge
Fish were randomly assigned to two groups, and transferred into one of two test tanks (200 l). Starry flounder were given an intraperitoneal injection of 0.1 ml
>
Real-time quantitative RT-PCR assay
NKCC2 expression levels were assessed by real-time quantitative PCR to examine the potential modulation of NKCC2 gene expression by the bacterial challenge. Preparation Total RNA (from anterior and posterior intestines and rectum) and the cDNA synthesis were performed as described above. The diluted cDNA template was subjected to PCR cycling with a LightCycler® 480 (Roche Diagnostics, Basel, Switzerland). The 205-bp NKCC2 gene fragment was quantified with the specific q-psNKCC2-F and q-psNKCC2-R primer pair (Table 1). Plasmid DNAs containing the amplified parts of the target mRNAs were prepared as standard samples. The PCR mixture consisted of 0.5 μM of each primer pair and the LightCycler® 480 SYBR Green I Master (Roche Diagnostics) in a final volume of 20 μl. The transcript copy number was calculated in reference to the parallel amplifications of known concentrations of the respective cloned PCR fragments. The NKCC2 transcript level in each sample was normalized against its own level of the 18S rRNA control (Kubista et al., 2006; Schmittgen and Livak, 2008). Triplicate independent assays per cDNA sample were performed.
Differences among samples were assessed by one-way analysis of variance, followed by Duncan’s multiple range test. All statistical analyses were performed using the SPSS ver. 10.0 software (SPSS, Inc., Chicago, IL, USA), and differences were considered significant at
>
Deduced amino acid sequence characteristics of starry flounder NKCC2 cDNA
The starry flounder NKCC2 cDNA (GenBank accession no. AB645956) was 4,137 bp in length and contained a single ORF of 3,129 bp encoding a polypeptide of 1,043 amino acids (aa). The NKCC2 5′-untranslated region (UTR) and 3′-UTR were 174 and 834 bp, respectively. The calculated molecular mass of the mature peptide was 114.6 kDa with a theoretical pI of 6.47. Multiple sequence alignments of starry flounder NKCC2 with other orthologs revealed that it shared comparably higher identities with piscine NKCC2s at the amino acid level (range, 59.5–88.4%; Table 2). According to the topology prediction, starry flounder NKCC2 contained 12 putative transmembrane domains and a long cytoplasmic C-terminal tail, all of which are characteristic of NKCC. There were four amino acid residues within transmembrane domain 4 that were conserved among NKCC family members (N248, R251, G254, and G268) (Fig. 1).
A phylogenic tree analysis was performed for the NKCC1 and/or NKCC2 sequences from starry flounder and other fish species and mammals using the neighbor-joining method (Fig. 2). The relationships revealed in the phylogenic tree agreed with known taxonomic appraisals. The starry flounder NKCC2 was located in the same branch as that of olive flounder and was closely related to that of tilapia and Japanese rice fish,
>
Tissue distribution of the flounder NKCC2 transcripts
Unstimulated starry flounder juveniles were used to investigate NKCC2 transcript levels in different tissues (Fig. 3). The NKCC2 transcript was detected at limited levels in the anterior and posterior intestines and rectum. NKCC2 basal expression levels in all intestinal segments were similar. NKCC2 was not found in the other tissues examined.
>
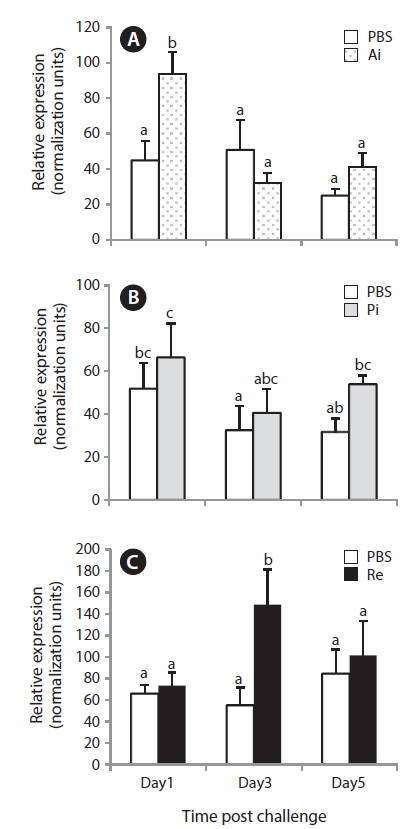
Expression of NKCC2 mRNA after S. parauberis challenge
The quantitative RT-PCR assay results revealed that the anterior intestinal NKCC2 transcript in the bacterial-challenged group increased significantly only 1 day post-challenge with
In the present study, we focused on the association between intestinal ion channel function and streptococcosis. We identified the cDNA encoding NKCC2 in the intestinal tract of starry flounder based on the deduced amino acid sequence and the comparably higher identities with other teleost fish NKCC2s. The starry flounder NKCC2 is a 1,043-aa protein with a proposed topology that features 12 putative transmembrane-spanning domains and predicted cytosolic amino and carboxyl termini. This structural prediction closely resembles those of other known NKCC2 proteins. In addition, the starry flounder NKCC2 contained highly conserved amino acid residues within transmembrane domain 4, which are important for the NKCC cotransporting function (Moreno et al., 2004; Gamba, 2005). These findings suggest that the starry flounder NKCC2 is a functional cation-Cl− cotransporter involved in ion transport in the intestinal tract.
Starry flounder NKCC2 transcripts were found only in the three intestinal segments (e.g., anterior and posterior intestines and rectum) with moderate expression levels under non-stimulated conditions. Other teleosts NKCC2s are expressed in several tissues, although distribution is not widespread. For example, NKCC2 is preferentially expressed in the intestine and kidney of the marine medaka,
In this study, only the rectal level of NKCC2 in starry flounder responded significantly to the bacterial challenge, indicating that excess salt may be transported into the rectum. This similar induction pattern was observed in olive flounder, in which the NKCC2 transcript is upregulated in the posterior intestine after