The family Bramidae in the order Perciformes comprises 7 genera and 22 species worldwide (Nelson, 2006), 6 genera and 10 species in Japan (Hatooka and Kai, 2013), and 4 genera and 5 species in Korea (Kim et al., 2005; Kim et al., 2012). The 5 species in Korea are Brama japonica Hilgendorf, 1878, Pteraclis aesticola (Jordan & Snyder, 1901), Pterycombus petersii (Hilgendorf, 1878), Taractichthys steindachneri (Döderlein, 1883), and Taractes asper Lowe, 1843. The genus Brama, family Bramidae, comprises 8 species worldwide (Mead, 1972; Pavlov, 1991; Moteki, 1995), but only one species, B. japonica, is recorded in Korea (Kim et al., 2012). However, the species of Brama are difficult to distinguish by external morphology (Mead, 1972). In December 2013, a single specimen of Brama was collected from Jeju Strait, Korea, and was identified as Brama orcini by both morphological and molecular methods. In this study, we describe the species and add it as a new member of the Korean fish fauna.
A single specimen of Brama orcini was collected by a purse seine in Jeju Strait, Korea on 9 December 2013. The following description was made on the basis of this specimen. Counts and measurements followed Hubbs and Lagler (2004) by a vernier caliper to the nearest 0.1 mm. The vertebrae were counted from a radiograph (Softex HA-100, Japan). The specimen was deposited at Pukyong National University (PKU), Korea. Molecular identification of this specimen was performed by a universal primer set VF2 (5´-TCAACCAACCACAAAGACATTGGCAC-3´) and FishR2 (5´-ACTTCAGGGTGACCGAAGAATCAGAA-3´) primers, which amplify the mitochondrial DNA (mtDNA) cytochrome c oxidase subunit I (COI) (Ward et al., 2005; Ivanova et al., 2007). Genomic DNA was extracted from muscle tissue using Chelex 100 resin (Bio-Rad, USA). A polymerase chain reaction (PCR) was performed in a total volume 50 μL containing DNA template 5 μL, dNTP 4 μL, 10X buffer 5 μL, Taq polymerase 0.5 μL, reverse primer 1 μL, forward primer 1 μL, and distilled water. PCR was conducted under the following conditions: initial de-naturation for 1 min at 95°C, followed by 35 cycles of 1 min at 95°C for denaturation, 1 min at 50°C for annealing, and 1 min at 72°C for extension, with a final extension at 72°C for 5 min. The PCR products were purified using ExoSAP-IT (Biochemical Co., USA). The PCR products were sequenced with the ABI PRISM BigDye Terminator v3.1 Ready Reaction Cycle Sequence Kit (Applied Biosystems, USA) on an ABI3730xi DNA Analyzer (Applied Biosystems). The sequence was aligned with ClustalW (Thompson et al., 1994) in BioEdit (ver. 7) (Hall, 1999). A neighbor-joining (NJ) tree (Saitou and Nei 1987) was constructed by the Kimura two-parameter model (Kimura, 1980) and in MEGA 5 (Tamura et al., 2011).
(New Korean name: Keun-bi-neul-sae-da-rae) (Fig. 1)
[fig.1.] Lateral view of three Brama species. (A1) Brama orcini, 267.0 mm standard length (SL), PKU 10115; (A2) Brama orcini, 188.3 mm SL, FAKU 132170; (B1) Brama japonica, 462.0 mm SL, PKU 562; (B2) Brama japonica, 446.0 mm SL, PKU 8590; (B3) Brama japonica, 198.6 mm SL, FAKU 131717; (B4) Brama japonica, 191.4 mm SL, FAKU 131718; (C1) Brama dussumieri, 100.1 mm SL, FAKU 133306; (C2) Brama dussumieri 137.5 mm SL, FAKU 134950.
Brama orcini Cuvier, 1831: 295 (type locality: eastern Indian Ocean); Mead, 1972: 72 (Indo-Pacific, East Pacific); Eschmeyer et al., 1983: 336 (USA); Masuda et al., 1984: 159 (Ryukyu Is. and Ogasawara Is., Japan); Smith, 1986: 634 (South Africa); Anderson et al., 1998: 24 (Maldive); Mundy, 2005: 375 (Hawaiian Archpelago); Hatooka and Kai, 2013: 908 (Sagami Bay, Ryukyu Is. and Ogasawara Is., Japan).
PKU10115, 267.0 mm standard length (SL), collected by a purse seine, Jeju Strait, (126°46′E, 33°53′N), 9 December 2013 (Fig. 1, A1).
Brama orcini: FAKU 132170, 188.3 mm SL, Sagami Bay, Japan (Fig. 1, A2), Brama japonica: PKU562, 395 mm SL, Wan-do, September, 2008 (Fig. 1, B1); PKU8590, 349.0 mm SL, Jeju Island, April, 2013 (Fig. 1, B2); FAKU 131717- 131719, 191.4-198.6 mm SL, Nohara, Maizuru, Japan (Fig. 1, B3-4), Brama dussumieri: FAKU 133306, 100.1 mm SL, Kawajiri-misaki, Yamaguchi, Japan, July, 2010 (Fig. 1, C1); FAKU 134950, 137.5 mm SL, Otomi, Fukui, Japan (Fig. 1, C2).
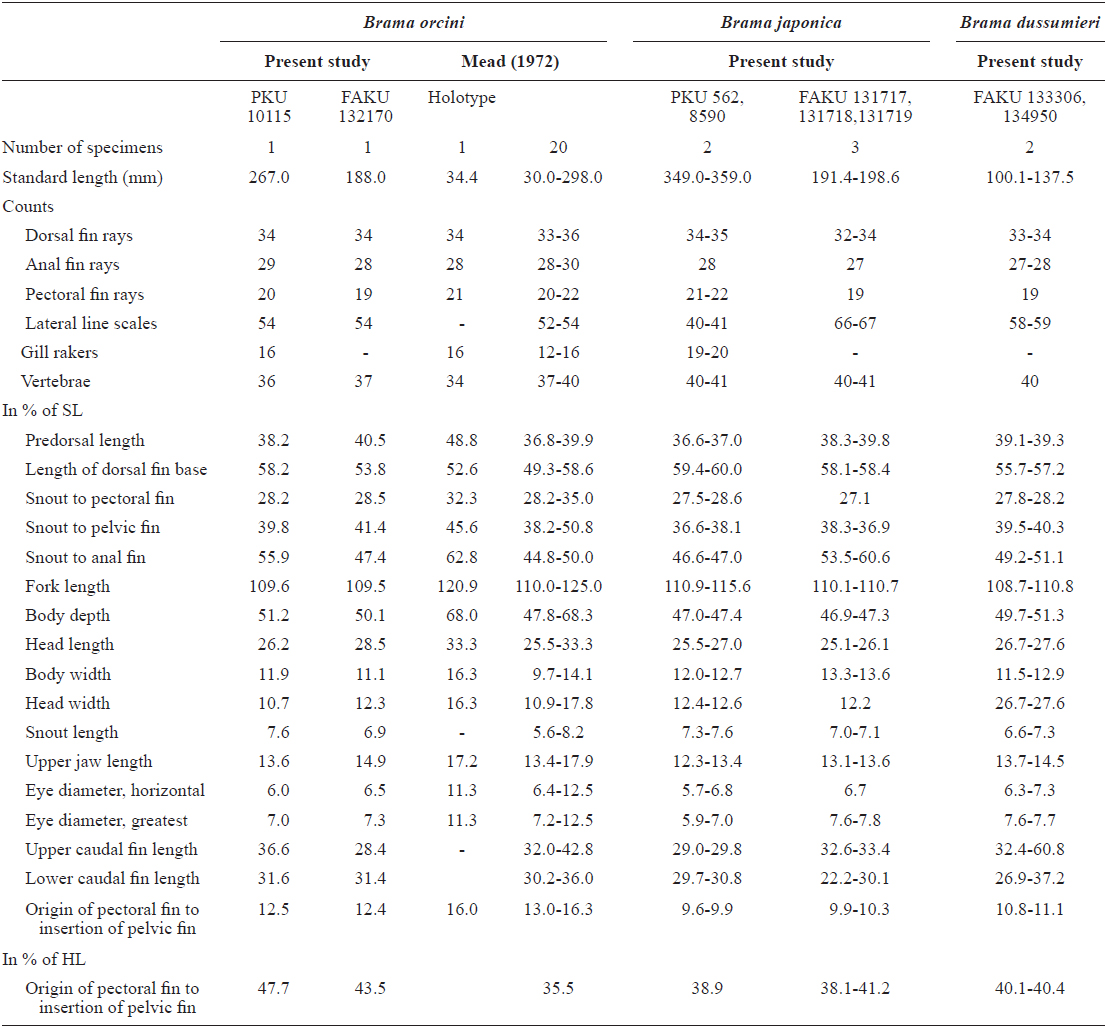
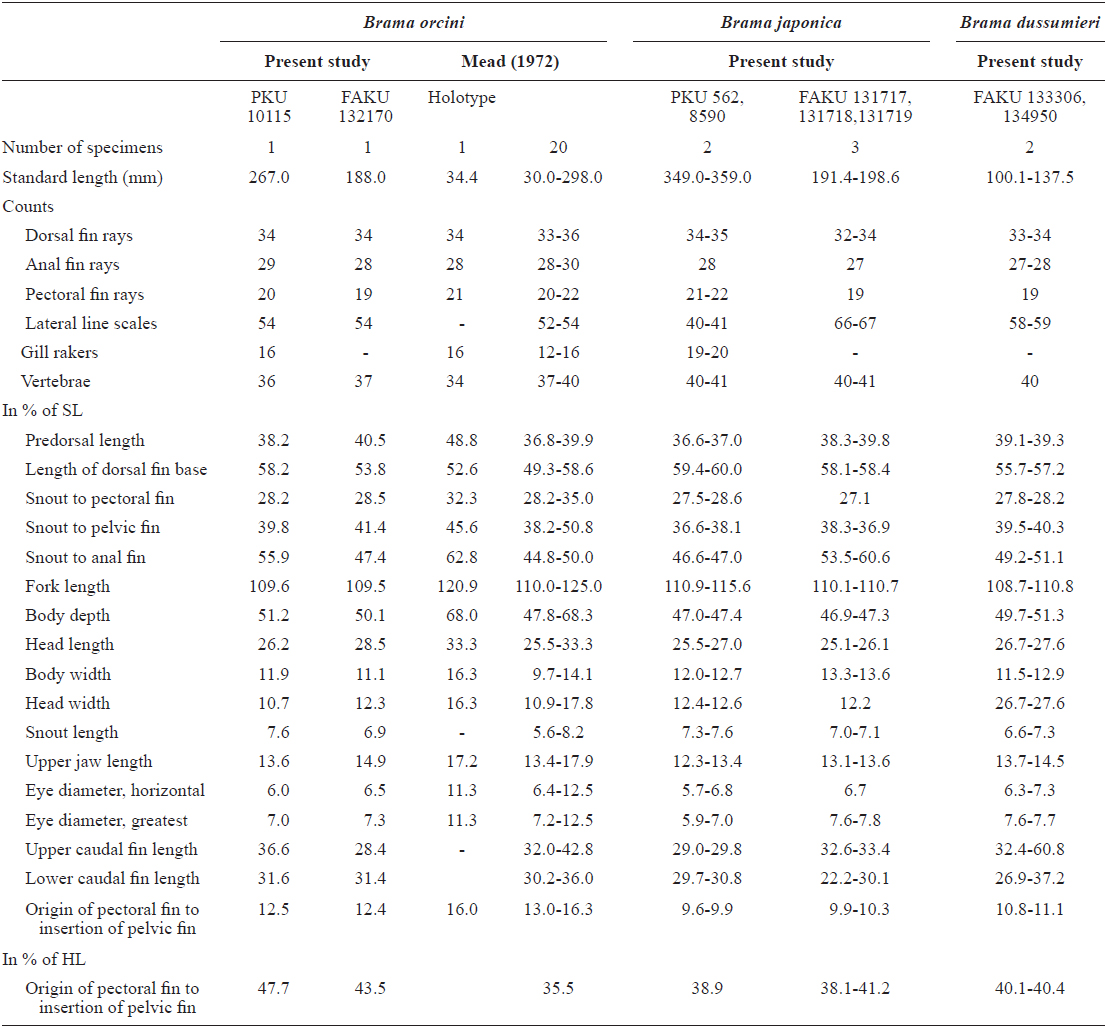
Dorsal fin rays 34; pectoral fin rays 20; anal fin rays 29; lateral line scales 54; gill rakers 16; vertebrae 36 (Table 1).
Body ovate, strongly compressed; body depth very high (51.2% in SL); head large, forehead slightly arched; snout short (7.6% in SL), slightly blunt; mouth strongly oblique, posterior margin of upper jaw extending slightly beyond line vertical to the middle of the pupil; upper jaw protruding more than lower jaw; one row of small conical teeth in the upper jaw, irregular rows of teeth in the low jaw, some mid section of teeth markedly large; eyes large (7.0% in SL), oblique and oval shaped; interorbital region convex; one pair of nostrils, round and located at anterior tip of snout; dorsal fin starts above posterior to origin of pectoral fin, end above the slightly anterior caudal peduncle; head and body completely covered with large oblong ctenoid scales; dorsal and anal fins covered with small scales; pectoral fin starts under edge of opercle and reaches to mid body; pelvic fin relatively short, and originating on vertical line posterior to the origin of the pectoral fin; caudal fin strongly forked, length of upper lobe (36.6% in SL) greater than that of lower lobe (31.6% in SL).
Color when fresh. Head and body silvery, but slightly darker dorsally; pectoral and pelvic fins translucent; dorsal and anal fin black.
Color after fixation. Head and body grayish yellow, but black dorsally; fin color same as when fresh.
Indo-Pacific, and East Pacific oceans, southeast Africa between 30ºN and 30ºS (Mead, 1972; Last and Moteki, 2001), Sagami Bay (Numazu, Shizuoka), Ryukyu Is. and Ogasawara Is., Japan (Hatooka and Kai, 2013) and Jeju Strait, Korea (present study).
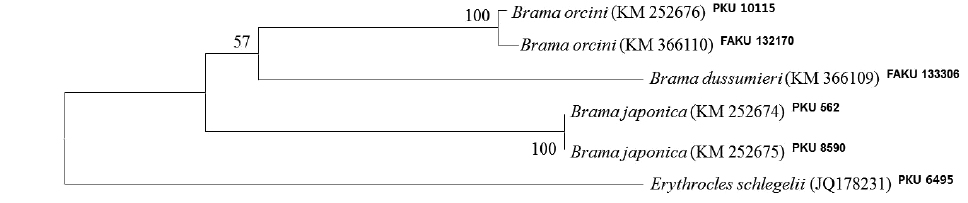
The present specimen collected from Jeju Strait, belongs to the genus Brama, dorsal fin originating behind head, over base of pectoral fin, the most of the elements in dorsal and anal fin branched, and both fins moderately stiff, erect and covered with ctenoid scales (Mead, 1972). The present specimen agreed well with the description of Mead (1972), who investigated the holotype and non-type specimens of B. orcini, in the numbers of lateral line scales (54), and gill rakers (16), and the distance between the origin of the pectoral fin base and the insertion of the pelvic fin base exceeding 12% SL. Other counts and measurements were consistent with those of a previous study of this species (Mead, 1972; Hatooka and Kai, 2013), except for the number of vertebrae: 36 in the present specimen vs. 38-40 in Mead (1972) vs. 37-40 in Hatooka and Kai (2013). The further study is required to reveal whether morphological difference between Korean and Japanese specimens of B.orcini is regional variations. Also, B. orcini is distinguished from the congeneric species B. japonica and B. dussumieri by the number of gill rakers (12-16 in B. orcini vs. 17-20 in B. japonica vs. 13-15 in B. dussumieri), the number of lateral line scales (52-54 in B. orcini vs. 65-75 in B. japonica vs. 57-65 in B. dussumieri), the distance between the origin of the pectoral fin base and the insertion of the pelvic fin base, expressed in terms of SL (greater than12% in B. orcini vs. less than 12% in B. japonica and B.dussumieri) and in head length (HL) (greater than 42% in B. orcini vs. less than 42% in B. japonica and B. dussumieri), and shape of the forehead arch (more strongly arched in B. japonica than in B. orcini) (Mead, 1972; Hatooka and Kai, 2013; Present study) (Fig. 1 A1-2, B1-4). To identify the species more accurately, we analyzed a total of 557 base pairs of the mtDNA COI sequence. The mtDNA COI sequence in the Korean B. orcini corresponded well with that of the Japanese B. orcini (genetic distance, d = 0.005), and differed considerably from that of the Korean B. japonica (d = 0.118) and the Japanese B. dussumieri (d = 0.120). Accordingly, we identified our specimen as B. orcini, based on both morphological and molecular characteristics. We propose the new Korean name “Keun-bi-neul-sae-da-rae” for B. orcini, because the species has relatively large scales as compared with those of congeneric species.