Pollutants with genotoxic potential are of great concern to many ecotoxicologists. Once released, these agents have the capability not only to cause morbidity and/or mortality in exposed organisms, but also to affect biological diversity at both intra- and interspecies levels (Atienzar et al., 1999). However, despite growing concerns over the presence of genotoxins in aquatic environments, adequately validated methods that can be used to evaluate genotoxicity and associated toxicity in aquatic organisms under environmentally relevant conditions are lacking. To examine genotoxic impacts, the three classic methods typically used are the micronucleus assay, the comet assay, and 8-oxoguanosine quantification. However, previous studies have demonstrated that in the eel,
Aroclor 1254, a commercial mixture of polychlorinated biphenyl (PCB) congeners, can be carcinogenic, clastogenic, and cause birth defects in both animals and humans (Cogliano, 1998; Sargent et al., 1989). While PCBs, such as dioxin, are usually considered to be non-genotoxic carcinogens (Whysner et al., 1998), there remains some uncertainly in this regard. The mechanisms by which PCBs exert their adverse effects are not fully understood and data on their genotoxic properties are controversial (Ross, 2004). Silberhorn et al. (1990) reported that Aroclor 1254 had negative results in the majority of genotoxicity assays. However, Aroclor 1254 produced DNA single-strand breaks in an alkaline elution/rat hepatocyte assay (Sina et al., 1983) and was clastogenic in dove embryos (Peakall et al., 1972). Accordingly, in this study, the genotoxic effect of Aroclor 1254 has been assessed using a random amplified polymorphic DNA (RAPD)-based methodology.
RAPD is currently used for intra-populational polymorphism detection and also enables the detection of genotoxicity after pollutant contamination of animals (Grayson et al., 1999, 2000). Moreover, Atienzar and Jha (2006) suggested that RAPD is a reliable, sensitive, and reproducible assay, and after proper optimization, has the potential to detect a wide range of DNA damage as well as mutations and therefore can be applied to genotoxicity and carcinogenesis studies. In fact, RAPD has been successfully used to detect ‘DNA effects’ induced by copper, cadmium benzo[
Some of the most persistent and ubiquitous contaminants are PCBs, although they are found only at low concentrations(Domingo and Bocio, 2007; Brown et al., 2006; Ross, 2004). Recently, the PCB contents of muscle tissues of marine fishes from nine coastal cities of East China were analyzed. The total PCB concentrations in all fish samples varied between 13.3 and 78.3 μg/kg lipid weight (lw), with an average of 35 ± 15 μg/kg lw (Xia et al., 2012). In our previous study on the toxicological effect of Arolcor 1254, the LC50 values in olive flounder embryos and larvae were 50.92 μg/L and 3.08 μg/L, respectively, and exposure to concentrations as low as 1 μg/L disrupted the early developmental stages of the fish (Min and Kang, 2013). An expert panel of the European Food Safety Authority (EFSA) concluded that we must improve our understanding of the environmental and human risks associated with PCBs due to their abundance in food and human tissues (Stenberg and Andersson, 2008).
According to Koh et al. (2001), the concentration of chlorinated compounds such as PCBs and dichlorodiphenyl trichloroethane (DDT) among the organic pollutants in Korean coastal environments was lower than in other developed countries. However, polycyclic aromatic hydrocarbons (PAHs), which are toxic compounds derived from oil spills, were present at higher concentrations in Korean coastal regions (Koh et al., 2001). Due to their high persistence and strong lipophilic properties, PCBs have bioaccumulated in aquatic food webs. Although the water solubility of Aroclor 1254 is only 2.7 μg/L, PCBs have been classified as probable human carcinogens by the Environmental Protection Agency (EPA), based on substantial evidence of cancer in animals (Walker, 1994). In fishes, PCBs such as Aroclor 1254 also are transported with lipoproteins into the yolk of developing oocytes (Ungerer and Thomas, 1996). Increased concentrations of PCBs in fish embryos and ovaries have been correlated with decreased viability, embryo toxicity, and embryonic malformations (Black et al., 1988; Weis and Weis, 1989; Min and Kang, 2013).
The development of such
For environmental monitoring purposes, one of the major biological challenges has been to bring potential organisms into continuous routine culture. This is especially important when sound genetic analysis is to be carried out. Olive flounder
The aims of this study were to detect potential genotoxic effects induced by Aroclor 1254 at environmentally relevant dosages using an RAPD technique, and to confirm the potential for the use of embryos and larvae of the olive flounder
>
Experimental animals and water conditions
Fertilized eggs of the olive flounder
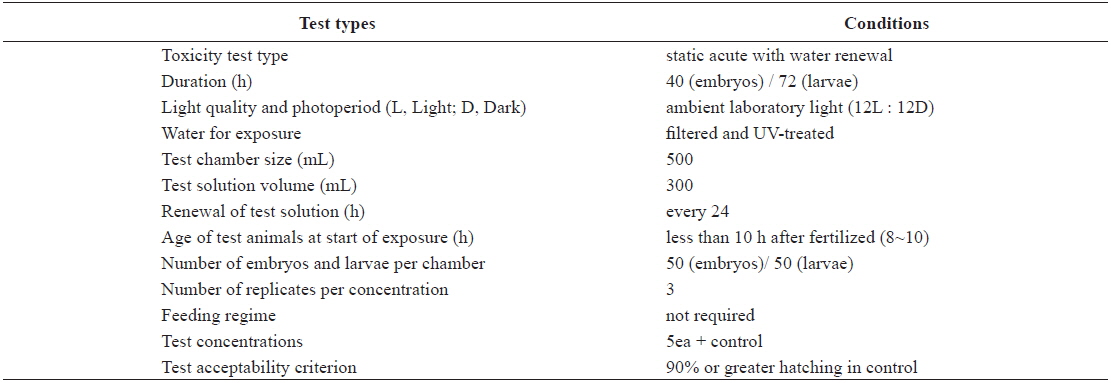
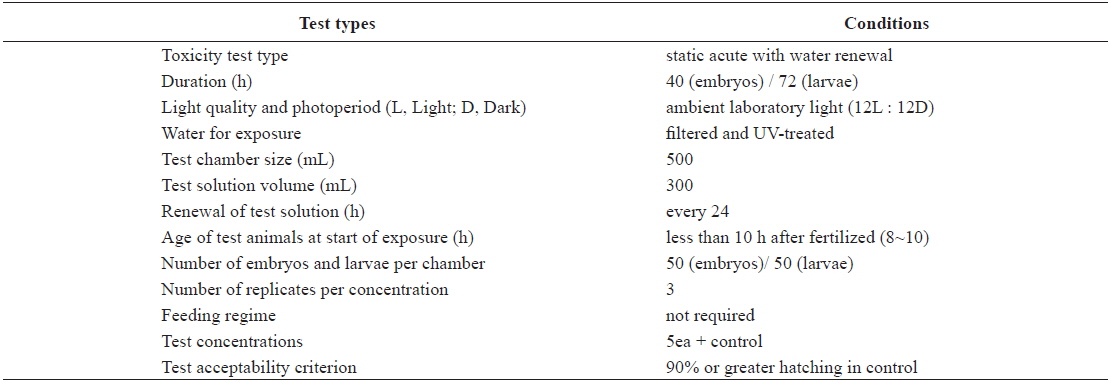
Test conditions for the acute toxicity tests with olive flounder Paralichthys olivaceus embryos and larvae
Aroclor 1254 was purchased from Dr. Ehrenstorfer GmbH (Augsburg, Germany). Five solutions (1, 5, 10, 20, and 40 μg/L) were prepared by diluting a stock solution. The stock solution was prepared with seawater that had been filtered through a grade GF/C Whatman filter (Maidstone, UK) and UV-treated.
>
Embryo and larval toxicity tests
The eggs obtained from the hatchery were approximately at the 8–10-h post-fertilization (blastula) stage. They were transferred directly into clean 500-mL glass beakers containing 0 (control), 1, 5, 10, 20, or 40 μg/L Aroclor 1254 for static exposure. Each Aroclor 1254 concentration (including the control) was carried out in triplicate beakers. Embryonic development was monitored every 3 h, until an endpoint at 40 h, or until hatching was complete. The beaker contents (Aroclor 1254 solutions) were replaced every 24 h throughout the toxicity test (Table 1). Newly hatched larvae were exposed to identical Aroclor 1254 concentrations to maintain the conditions (Table 1). The larvae were monitored every 6 h for up to 72 h.
At each time point during the embryo and larva toxicity tests, from each beaker, 50 organisms were collected, washed with UV-treated seawater, placed in phosphate-buffered saline (PBS), frozen in liquid nitrogen, and then stored at –70°C until subsequent analysis. The entire experiment was repeated in triplicate.
>
DNA isolation and DNA profiling using RAPD
The genomic DNA (gDNA) of exposed test organisms was extracted using a commercial DNA purification kit (NucloGen Biothechnology, Korea). Briefly, the organisms were lysed using sodium dodecyl sulfate and DNA was extracted by proteinase K digestion, phenol:chloroform extraction, and ethanol precipitation. DNA concentration was measured using a Bio-photometer (Eppendorf AG, Germany). DNA stock solutions were diluted to 10 μg/mL in distilled water and stored at –70°C until analysis.
DNA profiles of
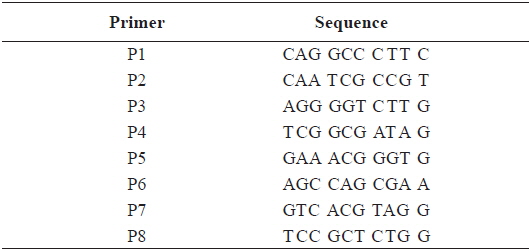
[Table 2.] Sequence of the primers used for RAPD PCR in this study
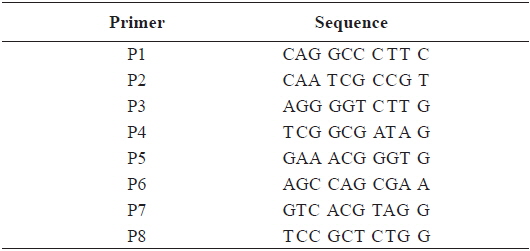
Sequence of the primers used for RAPD PCR in this study
Electrophoresis of RAPD reaction products were performed in 2% w/v agarose gels, using a Tris-Borate-EDTA buffer system (1X TBE; 90 mM Tris-base, 90 mM boric acid, and 2 mM EDTA) at 100 volts for 30 min. The gels were stained in a 1X TBE solution containing ethidium bromide (0.015% v/v) for at least 40 min. Gels were visualized under UV light, and photographed using a Gel-doc camera system (Gel-doc XR. Bio-Rad. USA). For comparison, a DNA molecular size marker (100-bp DNA Ladder, Bioneer, Co. Korea) was used.
The RAPD method is relatively new and there is no one standard statistical method used routinely for determining genetic diversity. For comparison, we used neighbor-joining analysis and frequency values (FV). Neighbor-joining analysis (Saitou and Nei, 1987), in which genetic distances are computed for all pairs of individuals, was performed using the NEIGHBOR program in PHYLIP 3.5c (Felenstein, 1993) to generate an unrooted phenogram. A phenogram was constructed based on all individuals examined in this study. In the phenogram, samples from the various concentration groups with similar genotypes can be identified through clustering. Also, in this study, the FVs obtained from calculating band counts of new appearances or disappearances (i.e., mismatched bands compared to the control) in the RAPD profiles using each primer were calculated as follows:
Frequency values of contamination indicative bands (FV) = (ΣNA + ΣND)/ NP ΣNA: the sum of the total number of newly appeared bands in each primer RAPD profile ΣND: the sum of the total number of disappeared band in each primer RAPD profile NP: the total number of primers used in the RAPD profile
The frequency values of contamination indicative profiles (FV) were calculated using a modification of a protocol outlined by Theodorakis and Shugart (1997). Treatment groups were compared to the control using a one- or two-way analysis of variance (ANOVA). If the ANOVA was significant (
The continuous presence of genotoxic chemicals in aquatic environments is of major concern due to their effects on the health of aquatic biota (Jha, 2004). DNA integrity is one of the biomarkers of pollution, and therefore much effort has been focused on finding rapid, sensitive methods for obtaining information on potential genotoxicity (Sarkar et al., 2006). In fact, several studies have used PCR-based techniques to assess genetic variation and changes in aquatic biota exposed to different contaminants (Theodorakis and Shugart, 1997; Nadig et al., 1998; Ma et al., 2000). In addition, it has been suggested that RAPD assays can detect mutations only if they occur in at least 2% of the DNA (John and Kortenkamp, 2000).
>
RAPD DNA profiles of embryos and larvae under Aroclor 1254 stress
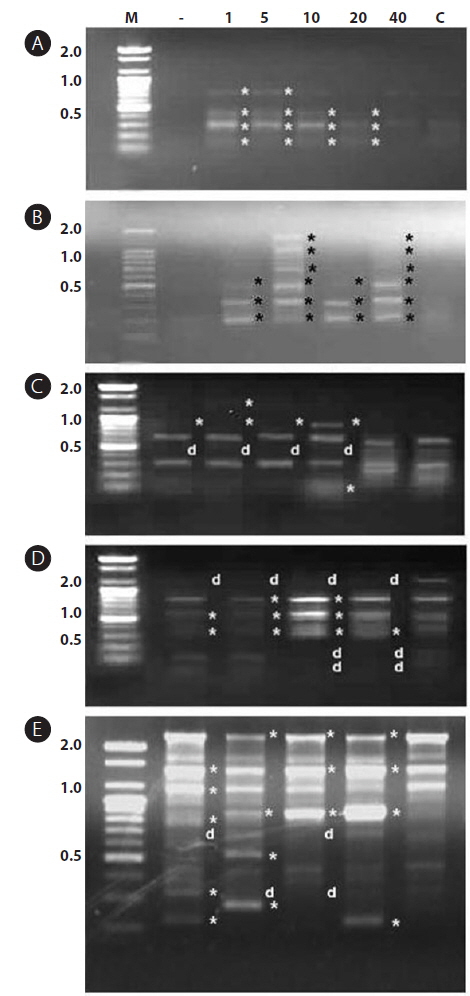
In this study, RAPD analyses were performed on gDNA pooled from 50 individuals of uncontaminated or Aroclor 1254-contaminated embryos or larvae. gDNA was pooled from 50 individuals to suppress intra-populational genetic polymorphisms that could potentially be revealed by RAPD. Eight different random primers were screened to discriminate controls from contaminated embryos and larvae. Of the eight primers tested, only four gave specific and stable results, and RAPD profiles generated with primers P2 and P6 were especially highly polymorphic. To determine the diversity of organisms exposed to Aroclor 1254, profiles were sorted based on the RAPD fragments at each exposure time (Fig. 1). The RAPD profiles showed substantial differences between unexposed and exposed animals, with apparent changes in the number, size, and intensity of amplified DNA fragments. Asterisks and the letter “d” on the RAPD profile gels of exposed groups indicate some of the obvious differences from the control (Fig. 1).
>
Analysis of data generated by RAPD profiling
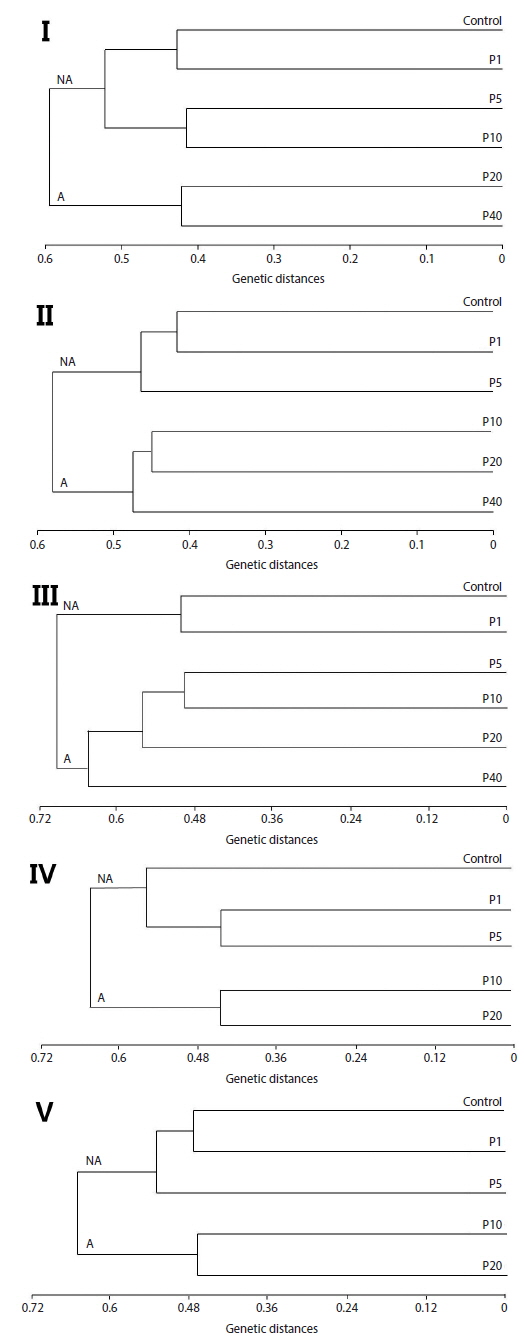
Neighbor-joining analysis assigns a genotype to each examined fish based on RAPD profiles generated by all analyzed primers and further reveals genetic relationships among all examined fish (Nadig et al., 1998). Because there was genetic differentiation among the treatment groups, a phenogram of genetic distance among the groups may be useful in delineating water quality (Fore et al., 1995). In this study, Fig. 2 is a phenogram generated using all primers and shows that the embryos were divides into two groups, designated here as the affected cluster (A), containing embryos affected by Aroclor 1254, and the non-affected cluster (NA). Interestingly, the number of the exposed group belonging to cluster (A) increased in an exposure-time-dependent manner in the embryo stage, increasing from two groups (20 and 40 μg/L) at 12 h, to four groups (5, 10, 20 and 40 μg/L) at 36 h (Fig. 2). In the larval stages, the phenogram also indicates two genetic clusters: cluster (NA) (1 and 5 μg/L), was more similar to the control than to cluster (A) (10 and 20 μg/L). The results demonstrate that the genotypes of embryos and larvae were highly variable, indicating that the genetic diversity of Aroclor 1254-contaminated groups can be readily analyzed by RAPD.
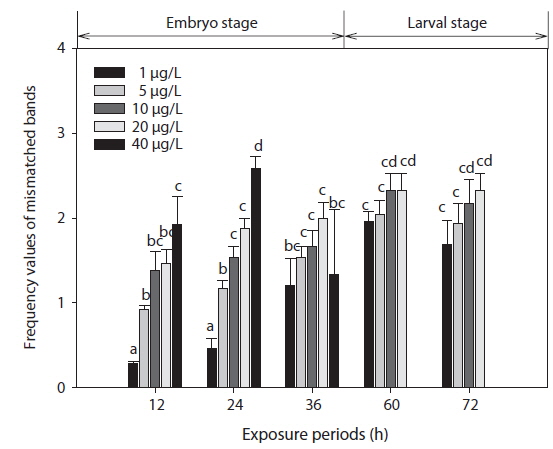
The phenetic numerical analysis of RAPD profiles is the most popular way to analyze RAPD in ecotoxicology studies and has been used to determine genetic diversity in nat-ural populations exposed to a variety of pollutants (Ross et al., 2002; Turuspekov et al., 2002). On a phenetic basis, the diversity is estimated by comparing the absence/presence of RAPD bands in exposed and non-exposed individuals (i.e., similarity analysis), or by counting their RAPD band numbers and considering band abundances (Maldonado et al., 2003). Modifications in band intensity and lost bands are likely to be due to one or a combination of the following events: (1) changes in oligonucleotide priming sites due to genomic rearrangements (more likely) and/or point mutations and DNA damage in the primer binding sites (less likely), and (2) interactions of DNA polymerase in organisms with damaged DNA (Liu et al., 2005). Thus, the FVs of these DNA polymorphisms could be used for the detection of genotoxic effects. In this study, changes in RAPD profiles were expressed as increases in FV (a measure reflecting the obvious changes in DNA patterns generated from toxicant-exposed organisms) in relation to profiles obtained from control organisms (Fig. 3).
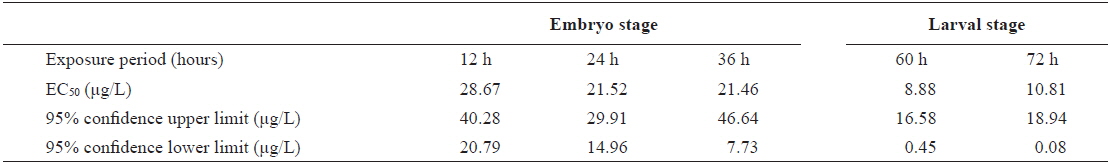
The EC50 values of FVs determined for embryos and larvae exposed to different concentrations of Aroclor 1254 are shown in Table 3. FVs significantly increased in a dose-dependent manner in developing embryos exposed to Aroclor 1254, but in the larval stage, there was no significant difference among the concentration groups (Fig. 3). Although FVs in larvae were not significantly different between the groups, a high FV was sustained until 72 h. This result suggests that during ELS, changes to gDNA stability may be an effect of the genotoxic potential of Aroclor 1254. This result also suggests that gDNA stability in the larval stage may be more sensitive than in the embryonic stage. As mentioned in our previous study (Min and Kang, 2013), our results demonstrate that Aroclor 1254 disrupted the early development stages of olive flounder, with higher toxicity in the larvae than in the developing, unhatched embryos.
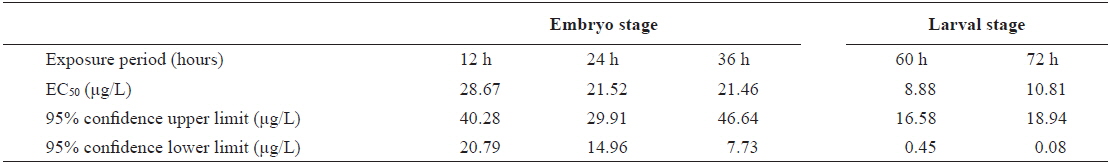
The value of EC50 of the frequency values (FV) determined by Probit analysis in olive flounder Paralichthys olivaceus embryos and larvae exposed to Aroclor 1254
Several studies have used ELS marine organisms to detect genomic instability caused by pollutants (Jha et al., 2000a, 2000b). Atienzar et al. (2002) suggested that 4-n-Nonylphenol and 17β-estradiol induced DNA effects in larvae of the barnacle
PCBs were not genotoxic in the bacterium
In the field of “genetic-ecotoxicology” or “eco-genotoxicology”, there are few well-validated tools for linking genetic damage to end points of direct importance to the ecological viability of marine populations (Anderson et al., 1994; Jha et al., 2000a). This study will provide a useful strategy in which first, RAPD analysis can be used as a screening method for potential genotoxic effects and, second, phenetic numerical analysis can be applied to measure DNA instability resulting from pollutants. These techniques are therefore being adopted as sensitive methods for the detection of induced genetic damage at the molecular level in both embryos and larvae of aquatic organisms.
In conclusion, our results indicate that Aroclor 1254 increased DNA damage in the embryo and larval stages (ELS) of olive flounder, as assessed by RAPD analysis. We suggest that DNA polymorphisms detected using RAPD analysis could be used for investigation of environmental toxicology and as a useful biomarker in an early warning system. In addition, we established this method under laboratory conditions using ELS and provided evidence of a reproducible and sensitive model for the detection of genotoxicants.