The genus Echinochloa (L.) P. Beauv. is a very difficult genus due to many interspecific and intraspecific forms found in the nature. This genus is still taxonomically controversial due to the absence of reliable morphological characters, resulting in poor taxonomic understanding of the genus. It has approximately 30- 40 species, mainly distributed in the tropical and warm temperate regions, usually associated with wet or damp places, of the world (Clayton and Renvoize, 1999).
This genus includes some of noxious weeds, such as E. crusgalli (L.) P. Beauv., E. oryzicola Vasinger, and E. colona (L.) Link in agricultural areas of the world. The dominance of these species causes serious yield loss if they are not appropriately controlled in the fields (Hill et al., 1985). The two species, E. crus-galli and E. oryzicola are problematic weeds in Korean agricultural fields as well. The Korean agronomists are confused about identifying this difficult genus using morphological characters because E. crus-galli not only exhibits very diverse morphological and ecological types, but also includes several varieties such as var. echinata (Willd.) A. Chev. and var. praticola Ohwi. However, these two varieties, echinata and praticola, have been treated as synonyms of var. crus-galli depending on taxonomists (The Plant List, 2014).
It has been known that eight morphological taxa of the genus, E. crus-galli var. crus-galli, var. echinata, var. praticola, E. oryzicola, E. colona, E. glabrescens Munro ex Hook.f., E. oryzoides (Ard.) Fritsch, and E. esculenta (A.Braun) H.Scholz, are distributed throughout Korea (Lee et al., 2013). Echinochloa crus-galli grows in the upland fields and also in paddy fields whereas E. oryzicola is frequently found in paddy fields. These two species are mainly found in the whole country. Existence of E. colona and E. glabrescens in Korea, however, should be reexamined. The result of a previous study showed that two species E. colona and E. glabrescens collected from Korea was clustered with E. crus-galli. They were not clustered with E. colona collected from China and/or formed independent clade (in press).
The continuous use of the herbicides which have the same mode of action has led to select the herbicide resistant species in the fields. Since the first herbicide resistant species, Monochoria vaginalis var. plantaginea (Roxb.) Solms, was discovered in Korea in 1998, the herbicide resistant E. crusgalli and E. oryzicola have also been reported and spreaded in Korea (Park et al., 2010). Since then, herbicide resistant Echinochloa spp. is getting more attention to both agronomists and farmers for effective control of these noxious weeds. Therefore, it is important to identify the species exactly and to know the genetic diversity of the species for designing the rational strategies for weed management, especially for the herbicide resistant weeds.
Simple sequence repeats (SSR) have many advantages over DNA sequencing for inferring phylogeny due to their neutral but faster evolution that may lead to more informative characteristics. Although the usefulness of SSR for resolving phylogenetic relationships among the closely related species was suggested (Takezaki and Nei, 1996), a few SSR phylogenies have been reconstructed (Petren et al., 1999; Ochieng et al., 2007). A better knowledge of the distribution of genetic variation within the Echinochloa would help its taxonomic classification and designing a rational strategy for herbicide test. The objective of this study was to identify and summarize the Echinochloa species distributed in Korea by comparing the genetic variation and relationship among Korean Echinochloa species using SSR fingerprinting.
total of 107 Echinochloa accessions was subjected to the fingerprinting study. Most of the Korean Echinochloa species were collected by authors from August to September of 2011 throughout the South Korea and the foreign accessions were the same accessions used for previous study (in press). The plant materials were identified based on the descriptions of Chen and Peterson (2006), Michael (2007) and Park et al. (2011). Plant materials are presented in Table 1.
A total genomic DNA was isolated from the green leaves of plant materials using a Genomic DNA Isolation Kit (NucleoGen, Germany) according to the instructions provided. The isolated DNA concentration and relative purity were checked using a Nanodrop ND-1000 (Dupont Agricultural Genomic Laboratory) and adjusted to 25 ng/μL for PCR amplification.
Five SSR markers developed by Danquah et al. (2002a) were subjected to study the genetic variation of the Korean Echinochloa species. Forward primers of the five SSR loci were labeled with blue (FAM), green (NED), or yellow (HEX) fluorescent tags (AB-PEC, Foster City, CA). PCR reaction mixtures were prepared according to Lee et al. (2005). The amplification was performed using a T100 Thermal Cycler (BIO-RAD, USA) that was set to run at 94°C for 3min for initial denaturation followed by 30 cycles of 94°C for 30 s, 55°C for 1min, 72°C for 90 s, and final extension at 72°C for 5 min. The amplification product of 1 μL was combined with 10 μL of Hi-Di formamide and 0.5 μL of an internal size standard, Genescan-500 ROX (6-carbon-rhodamine) molecular size standards (35-500 bp). The samples were denatured at 94°C for 3 min and analyzed with an ABI 3130xl Genetic Analyzer (Applied Biosystems / Hitachi, Inc., Foster City, CA, USA). Automated sizing and labeled alleles were determined and visualized relative to an internal Rox-labeled size standard using Genemapper 4.0 for automated data output.
Basic statistics at each SSR locus, including the number of alleles (NA), major allele frequency, gene diversity (GD), heterozygosity, and polymorphism information content (PIC), were calculated using the genetic analysis package PowerMarker ver. 3.25 (Liu and Muse, 2005). Genetic distances between each pair of the accessions were measured by calculating the shared allele frequencies using PowerMarker ver. 3.25 (Liu and Muse, 2005). The unweighted pair-group method with arithmetic averaging (UPGMA) was used to construct a phylogram from a distance matrix using MEGA ver. 5.2 (Tamura et al., 2011). The model-based clustering analysis was performed using all the 107 individuals and five independent runs of STUCTURE for each K value (the number of subpopulations) from two to 10 without prior population information. Runs were carried out by setting for 100,000 iterations with 100,000 burn-in and assuming an admixture model with correlated allele frequencies (Falush et al., 2003). For each K value, the runs showing the highest posterior probability of the data were considered. The true value of K was detected by an ad hoc quantity based on the second order rate of change of the likelihood function with respect to K (Evanno et al., 2005). An individual having more than 90% of its genome fraction value was assigned to a group.
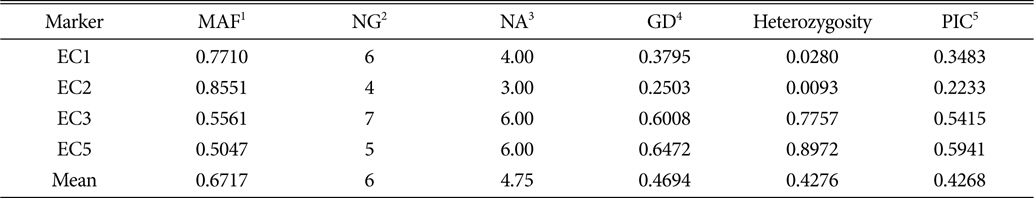
The 107 samples examined for SSR polymorphism represent seven species of Korean and foreign origins. The E. sp. in Table 1 were the taxa which were considered as either E. colona or E. glabrescens when they were collected. One of SSR markers, EC4, was not used to assess the genetic diversity of Echinochloa species because it did not show polymorphism across all the samples employed in this study. A total of 19 alleles, ranging from three (EC2) to six (EC3 and EC5), was detected from 107 Echinochloa accessions using four SSR markers with an average of 4.75 alleles per loci. The major allele frequency per locus varied from 0.5047 (EC5) to 0.8551 (EC2). The highest gene diversity (0.647) and Polymorphism Information Content (PIC; 0.594) were detected for the EC5, while the lowest gene diversity (0.250) and PIC (0.223) were detected using EC2 (Table 2). It was not congruent with the result of Danquah et al.(2002b) which revealed the least in EC1 (0.549) and the greatest in EC2 (0.715). Although this study included seven different species from several different origins, the average gene diversity, 0.469, seemed relatively lower than others comparing to those (0.556 and 0.66, respectively) of Danquah et al. (2002b) and Xu et al. (2004). The higher genetic diversity of Danquah et al. (2002b) might be due to inclusion of samples collected from broader geographic areas, such as the rice fields of Bangladesh, India, Côte D’Ivoire and Philippines. The low genetic diversity of the genus Echinochloa may mean that the genus will have not become a problem for agriculture in a certain changing environment. It could be, however, a big problem if they persist genes that can make the domesticated plants resistant to the intruder. Therefore, it is necessary to monitor the genetic diversity of this genus periodically in Korea.
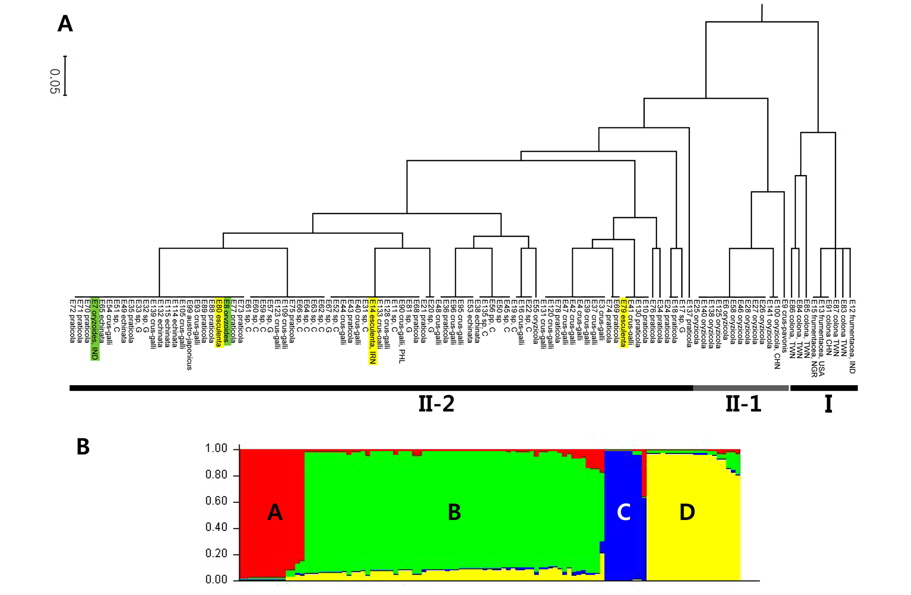
A genetic distance-based analysis was performed by calculating the shared allele frequencies among the 107 accessions and an unrooted and rooted UPGMA phylograms (Fig. 1A) were computed using MEGA5 software (Tamura et al., 2011). In the UPGMA phylogram, all Echinochloa accessions clustered into two main clades (I and II). The first group (I) included nine accessions which are E. colona, all coming from China and Taiwan, and its progenitor, E. frumentacea. Independent lineage of E. colona-frumentacea within the genus Echinochloa was also supported by many studies, including sequencing data derived from cpDNA and nrDNA in a previous study (in press) (Yamaguchi et al., 2005; Aoki and Yamaguchi, 2008). The second clade is composed of the remaining accessions, constituted by two clades (II-1 and II-2) again. II-1 is constituted by E. crus-pavonis and E. oryzicola. The rest of the accessions, a crus-galli complex, are arranged to the II-2 (E. crus-galli var. crus-galli, var. praticola, var. echinata, E. oryzoides, and E. esculenta). The close relationship between E. crus-pavonis and a crus-galli complex was supported by the maximum parsimony tree derived from cpDNA sequences, but not by that from nrDNA sequences (Aoki and Yamaguchi, 2008). We assume that the clustering in this clade II might be in relation to the number of chromosomes, 2n = 4× = 36 or 2n = 6× = 54; clade I is composed of the species having the number of chromosomes, 2n = 4× = 36, whereas clade II is consisted of the species with the hexaploids, 2n = 6× = 54. One interesting point is that E. oryzicola (E55) collected from Choonchon, Gangwondo, showed the close relationship with a crus-galli complex, but not with the other E. oryzicola accessions. Therefore, it is necessary to check the number of chromosomes of E. oryzicola (E55) collected from Choonchon, Gangwondo. If E55 has the number of chromosomes, 2n = 54, it will be a first report about the hexaploid E. oryzicola. Yamaguchi et al. (2005) called E. crus-galli, E. esculenta, and E. oryzoides as a crus-galli complex and considered to belong to the same species with more supporting studies such as isozyme analysis (Nakayama et al., 1999) and polymerase chain reactionrestiction fragment length polymorphism (PCR-RFLP) technique (Yasuda et al., 2002).
The model based population genetic structure of 107 Echinochloa individuals was conducted using STRUCTURE v2.3.3.(Pritchard et al., 2000). The true value of K was four and the real structure showed a clear peak of 107 Echinochloa individuals was set at K = 4 (Fig. 1B). What means that the 107 Echinochloa individuals should be divided into four subpopulations. In total, nine (C) and 14 (A) individuals were clearly assigned to two of the four subpopulations, whereas the rest of 84 individuals were divided into two different subpopulations (B and D). Subpopulations C and A were corresponded mainly to the clades I and II-1 of UPGMA dendrogram and subpopulations B and D were corresponded to the clade II-2.
[Fig. 1.] A. UPGMA dendrogram generated from 107 Echinochloa individuals using four SSR markers. Clade I was consisted of E. colona and its progenitor, E. frumentacea. Clade II was divided into two subclades, II-1 and II-2. Subclade II-1 was composed of mainly E. oryzicola and E. crus-pavonis whereas the rest the individuals, E. crus-galli, E. oryzoides, and E. esculenta, were arranged to the subclade II-2. B. Model-based populations of 107 Echinochloa individuals. Each accession is divided into a number of hypothetical sub-populations based on the proportional membership coefficients totaling 1 at K = 4. The two of subpopulations, C and A, were corresponded to the I and II-1 and the other two, B and D, were arranged to the clade II-2 of the UPGMA dendrogram.
The three clusterings, including subclade II-1 and II-2, of UPGMA phylogram might be more reasonable than four subgroups of the population structure because of no distinct morphological characters for each subpopulation found between the two subpopulations, B and D. Therefore, we have concluded from the results of the previous study (in press) and this study as follows: First, we could not find E. colona and E. glabrescens in Korea for our studies. Vouchers (KHB 1045294 and 1086605, KHB 1264632 and 1264634) loaned from Korea National Arboretum were turned out E. crus-galli, but neither E. glabrescens nor E. colona, respectively. Samples collected as those two species were E. crus-galli as well. Therefore, their natural distribution in Korea should be more surveyed. Second, Biological varieties praticola and echinata of E. crusgalli should be treated as E. crus-galli because many integrated forms can be easily found in the nature. In addition, some of the characters traditionally used for distinguishing taxa, such as awn length of var. echinata, are affected by the amount of moisture available in the habitats (Michael, 2007). Third, the Korean Echinochloa should be summarized with four species, i.e., E. oryzicola, E. crus-galli, E. esculenta, and E. oryzoides. The most abundant species is E. crus-galli. We also consider treating the latter three species, regarded as a crus-galli complex, as one species with care. Further detailed studies, however, should be warranted for better understanding of the Echinochloa in Korea.