RNA interference (RNAi), a highly conserved cellular mechanism in eukaryotic organisms, is a form of post-transcriptional homologous gene-silencing induced by double-stranded RNA (dsRNA) (Fire et al., 1998; Fire, 1999). DsRNA in cells is cleaved into small dsRNA fragments called the short interfering RNA (siRNA) by an RNase III-like enzyme, Dicer. These fragments are then incorporated into the RNA-induced silencing complex (RISC), which can degrade target mRNA in a sequence-specific manner, resulting in knock-down of the expression of the target gene (Bernstein et al., 2001; Sharp, 2001; Zamore, 2001; Hannon, 2002). Although RNAi-mediated knockdown of specific genes has been demonstrated in some species of gastropods, such as Aplysia (Lee et al., 2001), Lymnaea stagnalis (Korneev et al., 2002), and Biomphalaria glabrata (Jiang et al., 2006), and in bivalves such as the Pacific oyster Crassostrea gigas (Fabioux et al., 2009; Huvet et al., 2012) and Zhikong scallop Chlamys farreri (Wang et al., 2011), RNAi has not commonly been applied in mollusks.
Antimicrobial peptides (AMPs) are small amphipathic peptides that play a role in the innate defense of hosts against microbial infections (Hancock and Scott, 2000; Bulet et al., 2004; Otero-González et al., 2010). Big defensin, initially identified in the horseshoe crab Tachypleus tridentatus (Saito et al., 1995), is a type of AMP with two structurally and functionally different domains. The N-terminal domain is highly hydrophobic and more active against Gram-positive bacteria, while the C-terminal domain is cysteine-rich, cationic, and more active against Gram-negative bacteria (Saito et al., 1995). In bivalve mollusks, several big defensins have been reported. Zhao et al. (2007) identified the first one from bay scallop (Argopecten irradians) and showed that it had microbicidal activity against Gram-positive and Gram-negative bacteria and fungi. In the hard clam (Mercenaria mercenaria), expression of a big defensin gene significantly increased after challenge with a protistan parasite called quahog parasite unknown (Perrigault et al., 2009). A big defensin identified from clam (Venerupis philippinarum) showed increased transcription after Vibrio anguillarum challenge, and exhibited bactericidal activity against both Gram-positive and Gram-negative bacteria (Zhao et al., 2010). Recently, Gerdol et al. (2012) reported eight big defensins from the Mediterranean mussel (Mytilus galloprovincialis). In the Pacific oyster (Crassostrea gigas), six AMPs including three defensins (Cg-Def, Cg-Defh1, Cg-Defh2) and three big defensins (Cg-BigDef1, Cg-BigDef2, Cg-BigDef3) have been reported (Gueguen et al., 2006; Gonzalez et al., 2007; Rosa et al., 2011). Among the three big defensins, Cg-BigDef1 and Cg-BigDef2 were transcriptionally upregulated after injection with heat-killed or live bacteria (Rosa et al., 2011).
Although various methods can be used to analyze gene function, RNAi-mediated knock-down of gene expression is a convenient method for in vivo analysis of gene function. However, for in vivo experiments, it is important to understand whether the RNAi effect is systemic and how long the RNAi effect will last. In the present study, we investigated whether focal injection of a long dsRNA targeting Cg-BigDef2 could systemically induce RNAi-mediated transcriptional knock-down of C. gigas big defensin 1 and 2 genes over time.
Double-stranded RNAs (dsRNAs) specific to oyster big defensin 2 (Cg-BigDef2) and to green fluorescent protein (GFP) were synthesized in vitro using the Megascript RNAi kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. Briefly, DNA fragments partially encoding cDNA of Cg-BigDef2 and GFP genes were amplified via polymerase chain reaction (PCR) with gene-specific primers (Table 1) and cloned into the pGEM T-easy vector (Promega, Madison, WI, USA). After confirming the sequences, plasmids containing Cg-BigDef2 and GFP were digested with EcoRI/NcoI and EcoRI/HindIII, respectively. Then these genes were subcloned into the LITMUS 28i vector (New England Biolabs, Ipswich, 378MA, USA) and named pdT7-BigDef2 and pdT7-GFP, respectively. For single-stranded RNA (ssRNA) production, each vector was linearized with EcoRI and in vitro transcription was performed. Each antisense strand was synthesized after linearizing the vector with NcoI (Cg-BigDef2) or HindIII (GFP). Equal amounts of sense and antisense RNA were annealed to produce dsRNA. Digestion of the template DNA and any ssRNA remaining in the dsRNA, as well as purification of dsRNA, were performed using reagents in the Megascript kit (Ambion).
Pacific oysters (C. gigas; shell length 6–8 cm) were obtained from a commercial oyster farm in Korea and maintained in filtered seawater at 20°C. For the experiment, 36 oysters were randomly selected, divided into 3 groups of 12 oysters, put into a 250-L tank, and acclimated for 1 week without changing the water. After starting the injection experiment, cultured water in the tank was completely replaced once a day with newly filtered seawater. Oysters in each group were injected with dsRNA targeting Cg-BigDef2 or GFP into the adductor muscle at a dose of 50 μg per oyster. Oysters in the control group were injected with phosphate buffered saline (PBS) alone. At 12, 24, 48 and 72 h post-injection, three oysters in each group were randomly removed to analyze knock-down of target gene expression.
Hemolymph withdrawn from the adductor muscle was immediately centrifuged at 1,000 g for 20 min at 4°C to obtain hemocytes. A part of the adductor muscle, as well as the mantle and gill, were excised from each oyster. Total RNA was extracted using RNAiso plus reagent (Takara, Shiga, Japan). Prior to synthesizing first-strand cDNA, 1 μg total RNA was treated with RNase-free DNaseI (Promega). To synthesize first-strand cDNA, 1 μg total RNA treated with DNase was incubated with 0.5 μL random primer (0.5 μg/mL, Promega) at 80°C for 5 min and further incubated at 42°C for 60 min in reaction mixture containing 2 μL each of 10 mM dNTP mix (Takara), 0.5 μL M-MLV reverse transcriptase (Promega), and 0.25 μL RNase inhibitor (Promega) in a final reaction volume of 10 μL. PCR was performed with 2×Prime Taq Premix (Genet Bio, Daejeon, Korea), 1 μL 10-1 diluted cDNA template, and oligonucleotide primer pairs for amplification of the each target BigDef gene (Cg-BigDef 1, 2-F and Cg-BigDef 1-R or Cg-BigDef 2-R) and the control elongation factor (EF) gene (EF-F and EF-R) (Table 1). Thermal cycling conditions were 1 cycle of 3 min at 95°C (initial denaturation) followed by 35 cycles (for Cg-BigDef1), 27 cycles (for Cg-BigDef2), or 25 cycles (for EF) of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C, with a final extension step of 7 min at 72°C. PCR samples to be compared were electrophoresed on 1% agarose gels and stained with ethidium bromide (EtBr).
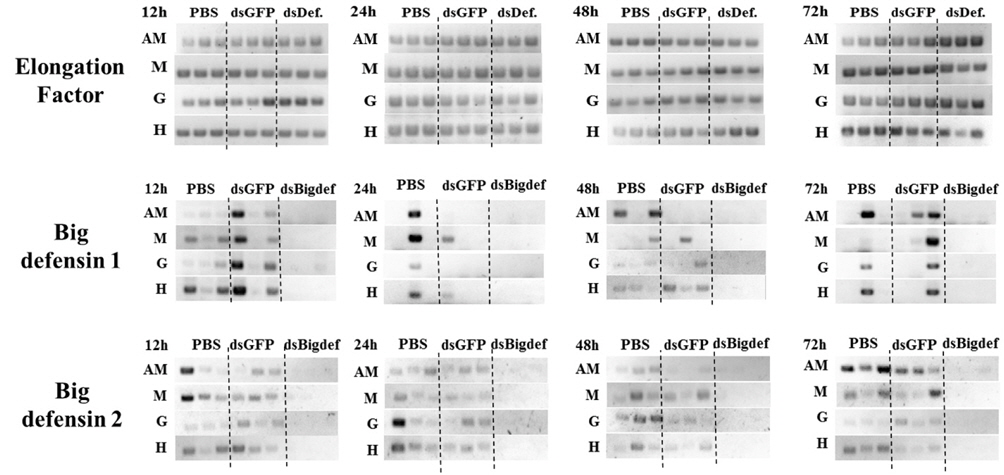
Transcription of the Cg-BigDef2 gene was clearly downregulated by injection of long dsRNA targeting the Cg-BigDef2 gene (Fig. 1). Transcription of the Cg-BigDef1 gene also decreased after injection of long dsRNA targeting the Cg-Big- Def2 gene. However, expression of the Cg-BigDef2 gene in oysters was not affected by injection of long dsRNA targeting the GFP gene.
Injection of long dsRNA targeting mRNA of the Cg-BigDef2 gene into the adductor muscle of oyster induced knockdown of the two BigDef genes in that muscle, as well as in the mantle, gill, and hemocytes (Fig. 1). Transcriptional knockdown of Cg-BigDef1 and Cg-BigDef2 genes in oysters injected with long dsRNA targeting the Cg-BigDef2 gene persisted until 72 h post-injection (Fig. 1).
Long dsRNA targeting mRNA of Cg-BigDef2 transcriptionally downregulated both Cg-BigDef1 and Cg-BigDef2. Because cDNA sequences of Cg-BigDef1 and Cg-BigDef2 are identical excluding an additional fragment of 20 nucleotides in Cg-BigDef1 (Rosa et al., 2011), transcription of both Cg-BigDef1 and Cg-BigDef2 may be suppressed by siRNAs originating from long dsRNA targeting Cg-BigDef2. Furthermore, we did not observe a decrease in expression of the two big defensin genes after injection of long dsRNA targeting GFP, suggesting that the knock-down of Cg-BigDef1 and Cg- BigDef2 was mediated by a sequence-specific RNAi phenomenon. Although Rosa et al. (2011) reported that hemocytes are the only tissues that express Cg-BigDefs, we found mRNAs of Cg-BigDef1 and 2 in hemocytes as well as other tissues, including the adductor muscle, mantle, and gill, which may be the result of hemocyte infiltration. Rosa et al. (2011) also indicated that other cell types can express Cg-BigDef genes.
In some organisms including Drosophila and vertebrates, dsRNA taken up by a cell is not transmitted to other cells (Roignant et al., 2003; Bitko et al., 2005), whereas in cells of Caenorhabditis elegans and some other invertebrates, dsRNA can spread to other cells, resulting in a systemic RNAi response (Winston et al., 2002; Dong and Friedrich, 2005). Fabioux et al. (2009) and Huvet et al. (2012) reported dsRNAmediated knock-down of target genes in oysters. However, the injection site and expression analysis in their experiments were restricted to the gonad, and little information is available on the use of systemic RNAi in oysters. In the present study, despite the injection of BD2-dsRNA into only the adductor muscle, knock-down of Cg-BigDef1 and 2 was observed not only in the muscle but also in hemocytes, the mantle, and gills, suggestive of systemic spread of RNAi in C. gigas.
The antimicrobial activity of molluscan big defensins has been demonstrated by analyzing in vitro bactericidal activity using recombinant big defensins or shown indirectly based on transcriptional upregulation of big defensin genes in response to bacterial infection (Zhao et al., 2007, 2010; Perrigault et al., 2009; Rosa et al., 2011). In the present study, we systemically downregulated two big defensin genes in C. gigas using RNAi, which can be used in future studies to analyze the functions of big defensins in oyster.