The family Pomacentridae (Perciformes) is one of the most diverse groups of reef fishes, and is distributed mainly in the tropical and temperate seas of the Indo-Pacific(Allen, 1991; Nelson, 2006; Allen and Erdmann, 2012). The Pomacentridae is comprised of approximately 350 species worldwide (Nelson, 2006), including 105 species in Japan(Aonuma et al., 2013), but only 17 species in Korea (Kim et al., 2005; Kim, 2011; Song et al., 2013). Although numerous studies have been conducted on the early life histories of pomacentrid species (Okiyama, 1988; Leis and Carson-Ewart, 2000; Kim et al., 2001; Richards, 2006; Murphy et al., 2007), identification of congeneric species remains difficult because of the extreme color variations that exist at all life stages (Neal, 1993; Song et al., 2013), and because research on only 15% of species has been conducted (Murphy et al., 2007). Because external morphological traits of larval and adult stages are very different, the characters used for adult stages cannot be used for larval stages (Blaxter, 1984; Miller and Kendall, 2009). Thus, molecular methods have been used widely in recent taxonomic studies of this group (Kim et al., 2008; Vandersea et al., 2008; Victor et al., 2009; Kwun and Kim, 2010; Ji et al., 2012; Kwun et al.,2012; Ko et al., 2013; Kwun et al., 2013; Lee and Kim, 2013).
In October 2010, a single juvenile pomacentrid specimen was collected from the Korea Strait (south of Tong-young), and based on molecular analyses, we identified it as a juvenile
A single juvenile pomacentrid specimen was collected from the Korea Strait (south of Tong-young) at a depth of 100 m by an RN80 net (Sanga, Busan, Korea) on 29 October 2010. The specimen was fixed in 99% ethanol immediately following collection. Counts and measurements were performed under an Olympus SZX-16 stereomicroscope (Olympus, Tokyo, Japan) following Leis and Carson-Ewart (2000). Measurements were recorded to the nearest 0.1 mm using an Active Measure image analyzer (Shinhan Scientific Optics, Seoul. Korea). Sketches of the external shape of the juvenile were created using a Olympus SZX-DA camera lucida (Olympus) attached to a microscope. Analyses of melanophore shapes and distributions followed Russell (1976). The specimen was deposited in the National Institute of Biological Resources (Voucher number, NIBR-P22498; GenBank accession number, JQ178234), Korea; other specimens are registered in Pukyong National University (PKU and PKUI), and were deposited in the collections of the Kanagawa Prefectural Museum of Natural History (KPM-NI).
Genomic DNA extraction and PCR analyses for molecular identification were performed according to Kwun and Kim (2010) and Kwun et al. (2012). The nucleotide sequences were deposited in the DNA Data Bank of Japan (DDBJ), in the European Molecular Biology Laboratory (EMBL), and in GenBank. Sequences were aligned using ClustalW (Thompson et al., 1994) in BioEdit version 7 (Hall, 1999). Sequence data were obtained for an adult
>
Chromis mirationis
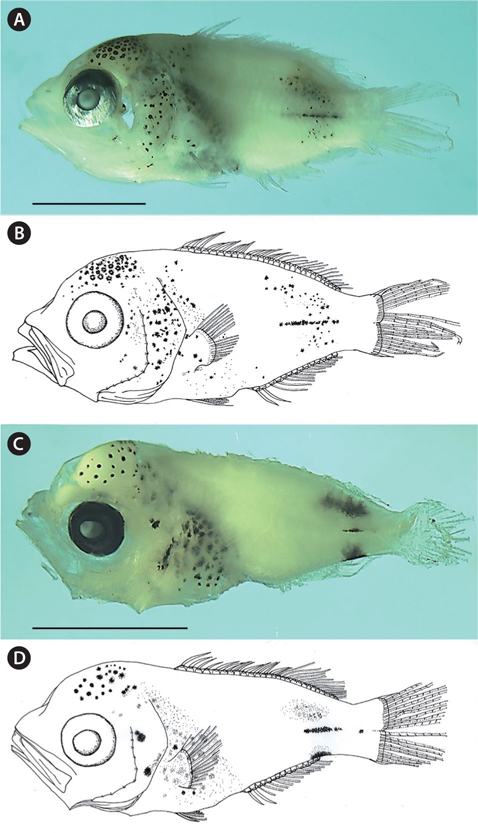
(New Korean name: Tti-ja-ri-dom; Fig. 1A and 1B)
NIBR P-22498, 5.9 mm standard length (SL), Korea Strait (south of Tong-young, 34˚20ʹ N, 128˚21ʹ E), 29 October 2010 (Fig. 1A and 1B).
>
Comparative material examined
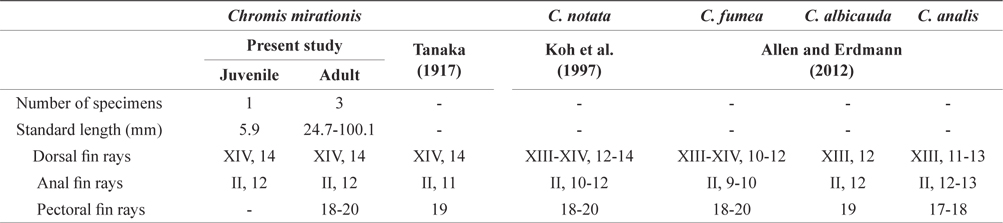
Counts are shown in Table 1. The following body measurements are expressed in terms of percent standard length (% SL): body depth, 44.2; head length, 46.6; snout length, 14.5; eye diameter, 16.9; upper jaw length, 26.0; predorsal length, 53.6; preanal length, 65.0; caudal peduncle depth, 16.0; caudal peduncle length, 11.7; dorsal fin base length, 49.2; anal fin base length, 33.7; pelvic fin length, 16.9; second anal fin spine length, 11.0.
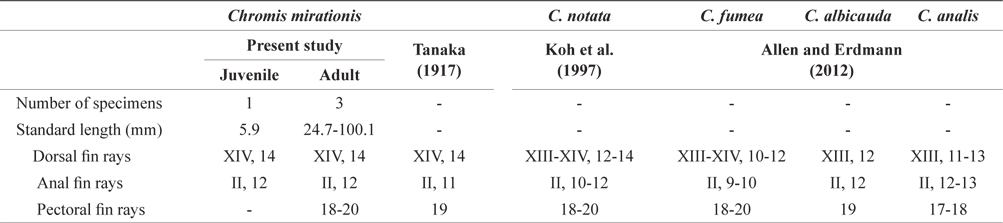
Comparison of meristic characters between Chromis mirationis and four species of genus Chromis
The body is short and slightly deep; the head and eyes are large, the snout is slightly pointed; the anterior margin of the head is slightly concave; the mouth is terminal, the posterior tip of the maxilla is located beyond the anterior margin of the eye; the teeth are small and conical on both jaws; the posterior margin of the preopercle is weekly serrated; the origin of the dorsal fin is located above the posterior margin of the opercle; the caudal peduncle is short; the anus is located behind the middle of the body; and the posterior tip of the pelvic fin reaches the anus.
Stellate melanophores are densely distributed on the occipital; punctate and punctate-stellate melanophores are scattered on the operculum, shoulder, abdomen, and pectoral fin base; rod-like stellate melanophores are sparsely distributed on the mediolateral; small punctate-stellate melanophores are scattered on the dorso- and ventro-laterals. Stellate melanophores are also densely distributed on the peritoneum. No melanophores exit on the snout, the middle region of the body, the dorsal and ventral contours, the caudal peduncle, the posterior end of the anal fin base, or the fin membranes (Fig. 1).
>
Mitochondrial DNA sequence analysis
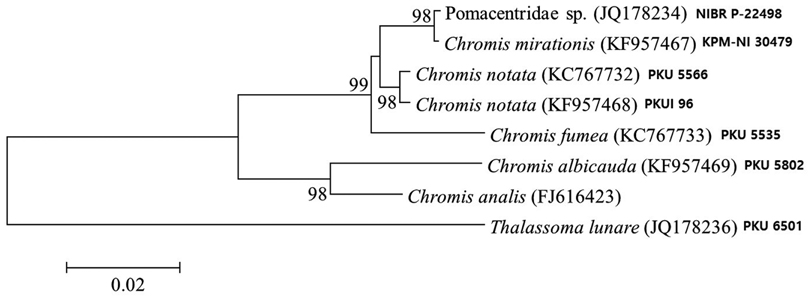
Analysis of 566 base pairs of mitochondrial DNA 16S rRNA sequences shows that our juvenile specimen is closely related to adult
The present juvenile specimen can be classified into the genus