Dinoflagellates are ubiquitous protists in marine environments (Jeong et al. 2010, Gomez 2012b), some forming red tides or harmful algal blooms that cause largescale mortality of fish (Deeds et al. 2002, Jeong et al. 2013). Dinoflagellates play various ecological roles in marine planktonic communities by serving as prey for diverse predators and acting as predators of diverse prey items (Jeong et al. 1999, 2013, Stoecker et al. 2006, Glibert et al. 2009, Hansen 2011, Sanders 2011, Lee et al. 2012, Kang et al. 2013, Park et al. 2013, Seong and Jeong 2013, Yoo et al. 2013). Identification or establishment of a new species is therefore a critical step in understanding the marine ecosystems, and elucidating the potentially harmful effects of dinoflagellates in the aquaculture industry.
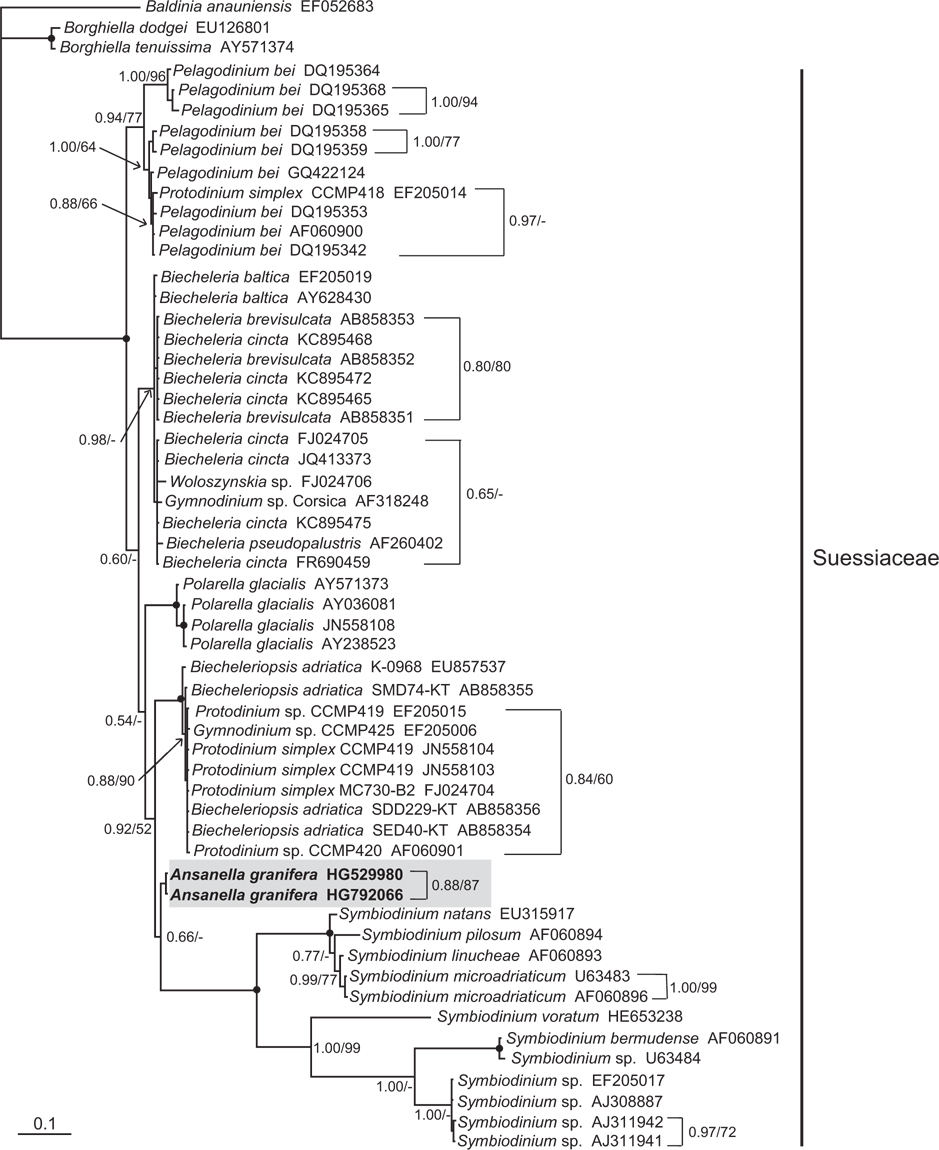
The order Suessiales was established by Fensome et al. (1993), with the two new families Suessiaceae and Symbiodiniaceae. Species in the family Suessiaceae comprised fossil dinosporin cysts with a 7-9 latitudinal paraplate series (i.e., arrangement of amphiesmal vesicles [AVs] in latitudinal series), whereas species in the family Symbiodiniaceae comprised extant symbionts with a 7-latitudinal plate series. However, the number of latitudinal series of AVs has since been extended to include species with more than 10 (Kremp et al. 2005). Moestrup et al. (2009a) merged Suessiaceae and Symbiodiniaceae, as the fossil species of the Suessiaceae are morphologically very similar to extant species such as Polarella (Montresor et al. 1999), and it seems unnecessary to establish two parallel taxonomic systems for extant and fossil species. Moestrup et al. (2009a) and Siano et al. (2010) included several genera in the family Suessiaceae, based on four major morphological characters: eyespot structure, morphology of the apical apparatus, structure of the resting cyst, and presence or absence of a nuclear fibrous connective, and also by molecular characterization. Siano et al. (2010) later proposed the number of latitudinal series of AVs as an additional morphological character for differentiating genera. Gomez (2012a) retained Symbiodiniaceae for the extant and Suessiaceae for the fossil species and included eight genera in this Symbiodiniaceae (Aureodinium Dodge, Biecheleria Moestrup, Lindberg and Daugbjerg, Biecheleriopsis Moestrup, Lindberg and Daugbjerg, Pelagodinium Siano, Montresor, Probert and de Vargas, Polarella Montresor, Procaccini and Stoecker, Piscinoodinium Lom, Protodinium Lohmann, and Symbiodinium Freudenthal). However, whether Aureodinium and Piscinoodinium belong here need to be supported by additional studies (Lohmann 1908, Freudenthal 1962, Dodge 1967, Lom 1981, Montresor et al. 1999, Moestrup et al. 2009a, 2009b, Siano et al. 2010, Gomez 2012a). Aureodinium has not been examined since its original description by Dodge (1967) and its phylogenetic affinities are uncertain. A molecular study (single subunit [SSU]) of the parasite Piscinoodinium, however, indicated phylogenetic relationship to the Suessiales (Levy et al. 2007), but additional information on amphiesmal plates and other morphological data are required to further evaluate the phylogenetic relationship.
Recently, we isolated thin-walled dinoflagellates from Shiwha Bay, Korea, and established two clonal cultures. The morphology of the cells from the cultures was almost identical, and there was only one base pair difference in the sequences of their SSU, internal transcribed spacer (ITS) region (ITS1, 5.8S, ITS2), and D1-D3 large subunit (LSU) rDNA. On the basis of morphological characters (in particular, the structure of the eyespot, thylakoids, and apical apparatus) and molecular analyses, these dinoflagellates are classified as a species of the family Suessiaceae, of which Symbiodiniaceae is considered to be a synonym. However, there is no other genus or species matching the characteristics of these dinoflagellates within the family. Therefore, in this study, we propose classifying these thin-walled dinoflagellates in the new genus Ansanella.
This study describes the morphological features of A. granifera gen. et sp. nov., using light microscopy, scanning electron microscopy (SEM), and transmission electron microscopy (TEM). The molecular sequences of the SSU, ITS region (ITS1, 5.8S, ITS2), and D1-D3 LSU rDNA from cultured cells are also described, as well as pigment data obtained using high-performance liquid chromatography (HPLC).
Samples of surface sediment were collected from Shiwha Bay, Korea (37˚18′ N, 126˚36′ E) in 2010 and 2012, using an Ekman grab (Wildco; Wildlife Supply Company, Buffalo, NY, USA). Samples were stored in the dark at 4℃ until processed further. The sediments were sieved consecutively through 100-µm and 15-µm Nitex meshes. The material on the 15-µm mesh was then transferred to a 50-mL beaker containing filtered seawater. A manual vortex was applied and the suspended sediment fraction was recovered. The sediment fraction was incubated in f/2 medium without Si (Guillard and Ryther 1962), in a growth chamber at 20℃ under 20 µmol m-2s-1 illumination, in a 14-h light, 10-h dark cycle. The incubated sediments were observed regularly for the presence of motile cells, using a stereomicroscope (SZX-12; Olympus, Tokyo, Japan).
In 2013, one clonal culture of A. granifera from sediments collected in September 2010 (A. granifera AGSW10) was established using two serial single-cell isolations, and another clonal culture was established from sediments collected in December 2012 (A. granifera AGSW12). The temperature and salinity of the ambient waters during collection of the sediments were 21.3℃ and 15.6, respectively, in 2010, and 2.0℃ and 28.0, respectively, in 2012. When the concentration of A. granifera had increased sufficiently, aliquots of cells were transferred to 32-mL, 270-mL, and 500-mL polycarbonate bottles containing fresh f/2 medium. The bottles were placed on a shelf, and incubated under the same conditions as those used for the sediments.
The morphology of A. granifera was examined using light microscopy, SEM, and TEM. An image analysis system was used to measure the length and width of live vegetative flagellated cells in images captured using a compound microscope (Zeiss Axiovert 200M; Carl Zeiss Ltd., Gottingen, Germany).
For SEM, a 20-mL aliquot of a dense culture of A. granifera was fixed for 10 min in 1% osmium tetroxide (final concentration). The fixed cells were collected on a 3-μm pore size, polycarbonate membrane filter without using additional pressure, and rinsed three times with distilled water to remove the salt. The filters with attached cells were dehydrated using a graded ethanol series (10, 30, 50, 70, 90, and 100% ethanol), followed by two changes of 100% ethanol, and then dried using a critical point dryer (CPD 300; Bal-Tec, Balzers, Liechtenstein, Germany). The dried filters were mounted on a stub and coated with gold-palladium in a sputter coater (SCD 005; Bal-Tec). Cells were viewed with a field-emission scanning electron microscope (S-4800; Hitachi, Hitachinaka, Japan).
For TEM, cells from a dense culture were transferred to a 10-mL tube, and fixed in 2.5% glutaraldehyde (final concentration). After 1.5-2 h, the contents of the tube were placed in a 10-mL centrifuge tube, and concentrated at 1,610 ×g for 10 min in a Vision Centrifuge VS-5500 (Vision Scientific Company, Bucheon, Korea). The pellet from the tube was subsequently transferred to a 1.5-mL tube and rinsed in 0.2 M sodium cacodylate buffer at pH 7.4. After several rinses, the cells were post-fixed for 90 min in 1% osmium tetroxide (final concentration) in deionized water. The pellet was then embedded in agar. Dehydration was performed using a graded ethanol series (50, 60, 70, 80, 90, and 100% ethanol), followed by two changes of 100% ethanol. The material was embedded in Spurr’s low-viscosity resin (Electron Microscopy Sciences, Fort Washington, PA, USA) (Spurr 1969). Sections were prepared using an RMC MT-XL ultramicrotome (Boeckeler Instruments Inc., Tucson, AZ, USA) and stained with 3% aqueous uranyl acetate followed by lead citrate. The sections were viewed with a JEOL-1010 transmission electron microscope (JEOL Ltd., Tokyo, Japan).
Three to five cells of A. granifera were transferred to a 1.5-mL tube containing 10-µL distilled water. The tube was then stored at -72℃ for 1-3 min to completely disrupt the cells. DNA extraction, amplification of the SSU, ITS1, 5.8S, ITS2, and D1-D3 LSU rDNA regions, PCR reactions, sequencing, and alignment were performed according to protocols used previously by Kang et al. (2011a).
Phylogenetic analyses of the SSU and LSU rDNA regions of A. granifera were conducted using MEGA v.4 (Tamura et al. 2007) and Clustal X2 (Larkin et al. 2007), including sequences obtained from GenBank. Maximum-likelihood (ML) analysis of the two regions was conducted using the RAxML 7.0.3 program (Stamatakis 2006), with a default GTR + G + I model. Tree likelihoods were estimated using a heuristic search with 100 random additional sequence replicates, and tree bisection and reconnection branch swapping. ML bootstrap was also conducted. Bayesian analyses were performed using MrBayes v.3.1 (Huelsenbeck and Ronquist 2001, Ronquist and Huelsenbeck 2003) with the default GTR + G + I mode, to determine the best available model for the data of each region. For all sequence regions, four independent Markov Chain Monte Carlo runs were performed according to methods used previously by Kang et al. (2010).
Approximately 100,000 cells of A. granifera were transferred to a 50-mL tube. The 50-mL aliquot was filtered onto a 1.2-µm pore-sized GF/C filter. A total of 3 mL of 95% methanol was used for extraction, and a Waters C8 column (150 mm × 4.6 mm, 3.5-µm particle size, 0.01-µm pore size) was used for separation. Analysis of pigments was performed using HPLC (LC-10A system; Shimadzu Co., Kyoto, Japan) according to methods used previously by Zapata et al. (2000). Pigments were identified and quantified by comparing retention times and absorption spectra with those of authentic standards. All standards were purchased from DHI Water and Environment (Horsholm, Denmark).
Phylum Dinoflagellata Butschli 1885 sensu Fensome et al. 1993
Class Dinophyceae Pascher 1914
Order Suessiales Fensome et al. 1993
Family Suessiaceae Fensome et al. 1993
Description. Free-living dinoflagellates with very thin transparent polygonal AVs arranged in longitudinal rows. The AVs are further arranged in 10-14 horizontal rows: 4-6 rows on the episome, 3-4 rows in the cingulum, and 3-4 rows on the hyposome. A single straight elongated apical vesicle, containing a single row of globular knobs, is present on the cell apex. Grana-like thylakoids in the chloroplasts and an eyespot of Type E are present. The peridinin-containing chloroplasts are lined by three membranes. A 51-base pair fragment of domain D2 of the large subunit rDNA, which is not found in genus Biecheleria, is present. Nuclear fibrous connective, nuclear envelope chambers, rhizocysts and lamellar body are absent.
Type species. A. granifera H. J. Jeong, S. H. Jang, Moestrup and N. S. Kang.
Etymology. The generic name “ansanella” refers to the name of the city, Ansan, enclosing Shiwha Bay where this species was collected.
Description. Episome conical with a round apex, larger than the trapezoidal hyposome. The cell has a wide and distinctive cingulum, which is displaced about its own width. The length and width of living cells are 10-15 µm and 9-12 µm, respectively. The ratio of cell length to cell width of living cells is 1.1-1.3. The nucleus is oval and located in the anterior to central part of the cell. Chloroplasts are yellowish-brown with the lobes arranged mainly near the surface of the cell; three or four starchenveloped pyrenoids are penetrated by thylakoids; pusule present. Peduncle and trichocysts absent. Estuarine and marine dinoflagellate.
Holotype. A holotype slide labeled USNM slide 1231539, of a culture fixed in 1% (final concentration) osmium tetroxide, was deposited in the Protist Type Specimen Slide Collection, US Natural History Museum, Smithsonian Institution, Washington, DC, USA.
Molecular characterization of Ansanella granifera: The rDNA gene sequence (SSU, ITS1-5.8S-ITS2, and D1-D3 LSU rDNA), GenBank accession Nos. HG529978-HG529980 (3,663-bp product) and HG792066 (3,663-bp product).
Type locality. Shiwha Bay, Korea (37˚18ʹ N, 126˚ 36ʹ E).
Etymology. The specific name “granifera”, indicates the characteristic chloroplast grana-like structures of this organism.
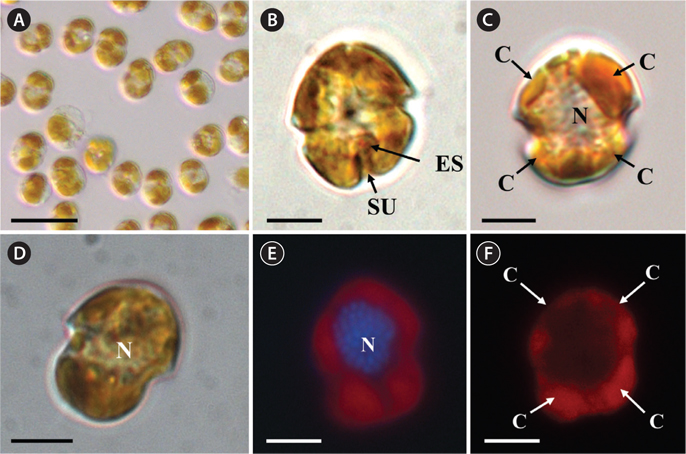
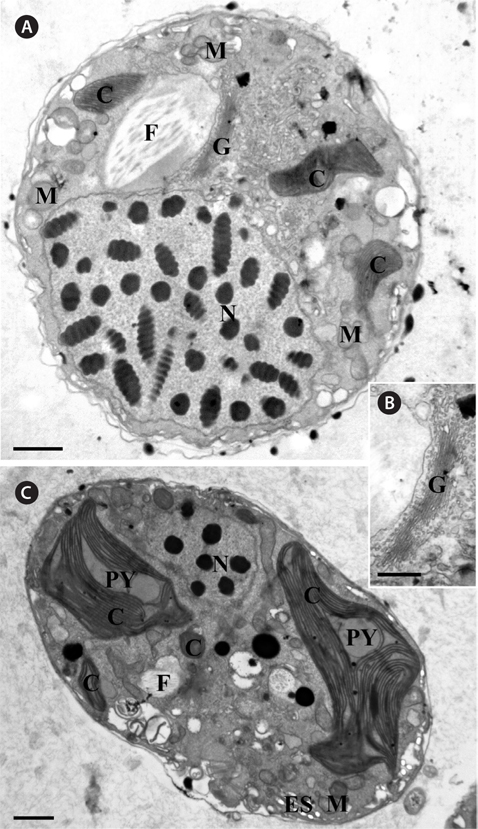
The morphologies of all the observed cells of Ansanella granifera AGSW10 and Ansanella granifera AGSW12 were almost identical. The episome was conical with a round apex, and was larger than the trapezoidal hyposome (Fig. 1A-F, Appendix 1A-C). There was a distinct elongated apical vesicle (EAV) (Figs 2A, C, 3C, D, 4A & C, Appendix 1F). The cingulum was displaced about its own width (Figs 2A & 4A, Appendix 1A). The sulcus became wider toward the antapex (Figs 1B, 2A, 3F & 4A, Appendix 1A). The nucleus was oval and located in the anterior to central part of the cell (Fig. 1C-E, Appendix 1B & C). Five to eight yellowishbrown chloroplasts were arranged in bands near the cell periphery (Fig. 1C & F, Appendix 1B). A bright red eyespot was located ventrally near the sulcus (Fig. 1B, Appendix 1A). One to three round pyrenoids were often visible in the light microscope (data not shown). The ranges (mean ± standard error, n = 40) of the length and width of living cells were 10.0-15.0 µm (12.6 ± 0.2 µm) and 8.5-12.4 µm (10.3 ± 0.2 µm), respectively. The ratio of cell length to cell width was 1.1-1.3 (1.2 ± 0.01 µm).
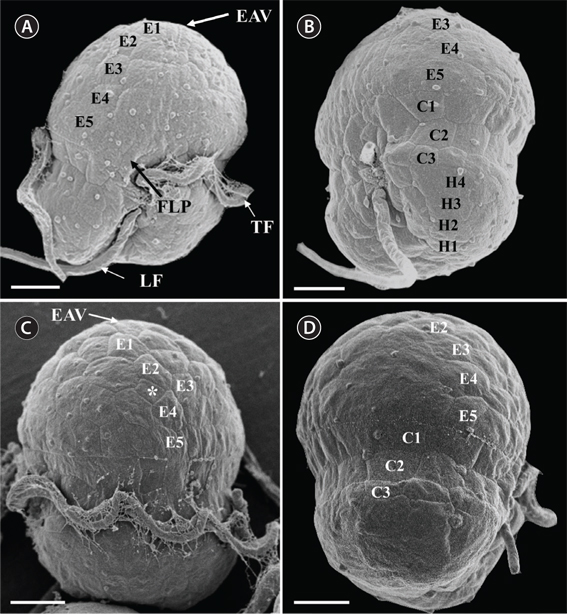
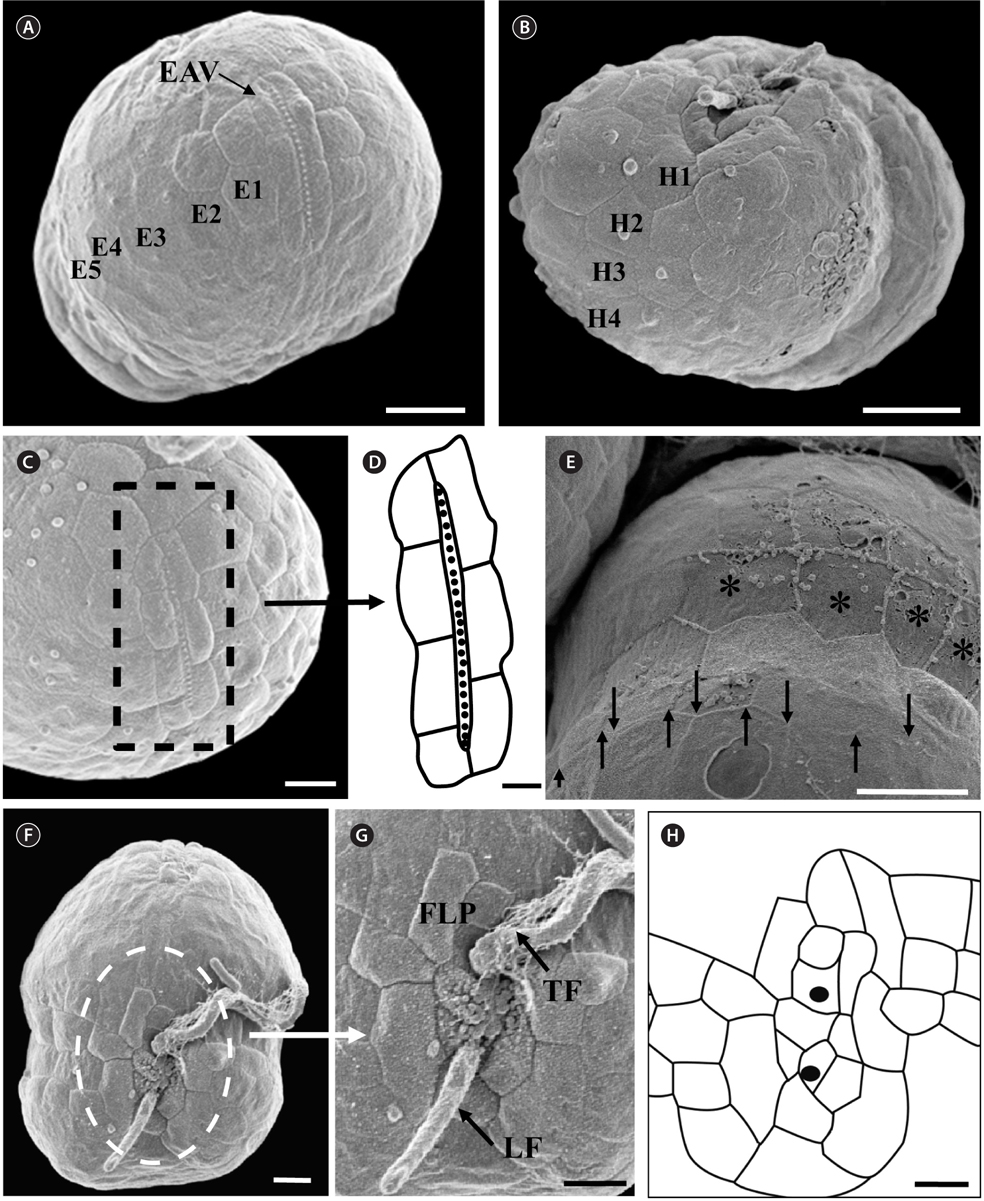
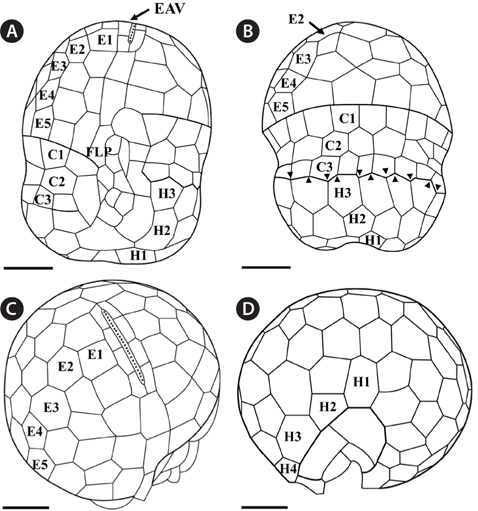
Under the SEM, the elliptical to round episome appeared larger than the trapezoidal hyposome (Fig. 2A-C). Cells were covered with hexagonal or pentagonal AVs. The length of each side of a hexagonal or pentagonal AV was 1.1-2.5 µm. The AVs were arranged in 10-14 rows (mostly 11, n = 100), as follows: 4-6 rows on the episome (mostly 5, n = 100), 3-4 rows in the cingulum (mostly 3, n = 100), and 3-4 rows on the hyposome (mostly 3, n = 100) (Figs 2A-D, 3A, B & 4A-D, Appendix 1D & E). The total number of AVs was ~160. The apical furrow apparatus consisted of a narrow EAV, approximately 4.1 µm, which extended from the mid-ventral side of the episome, over the apex to the dorsal side (Figs 3A, C, D & 4C). The EAV was ornamented with a central row of approximately 20-25 small knobs (Figs 3A, C, D & 4C), and was bordered by X+4-6 platelets (Figs 3A, C, D & 4C, Appendix 1F & G). In the dorsal part of Ansanella granifera cells, a small AV appeared to be constantly present (asterisk in Fig. 2C). The fingerlike protrusion on the right side of the episome extended to the intercingular-sulcal region (Figs 2A, 3G & 4A). On the dorsal side, vesicles of the third cingular row were hexagonal or pentagonal, forming a zigzag line along the lower cingular margin (arrows in Fig. 3E). In the sulcal region, two flagella emerged from flagellar pores, but no peduncle was observed (Fig. 3F-H). When the external membranes of the cell had disappeared, the contents of the AVs became very distinct (asterisk in Fig. 3E). The sulcus contained 6-7 rows of AVs, and became wider toward the antapex (Figs 3F-H & 4A).
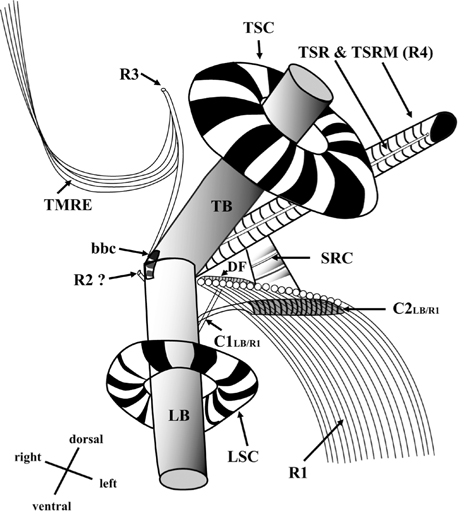
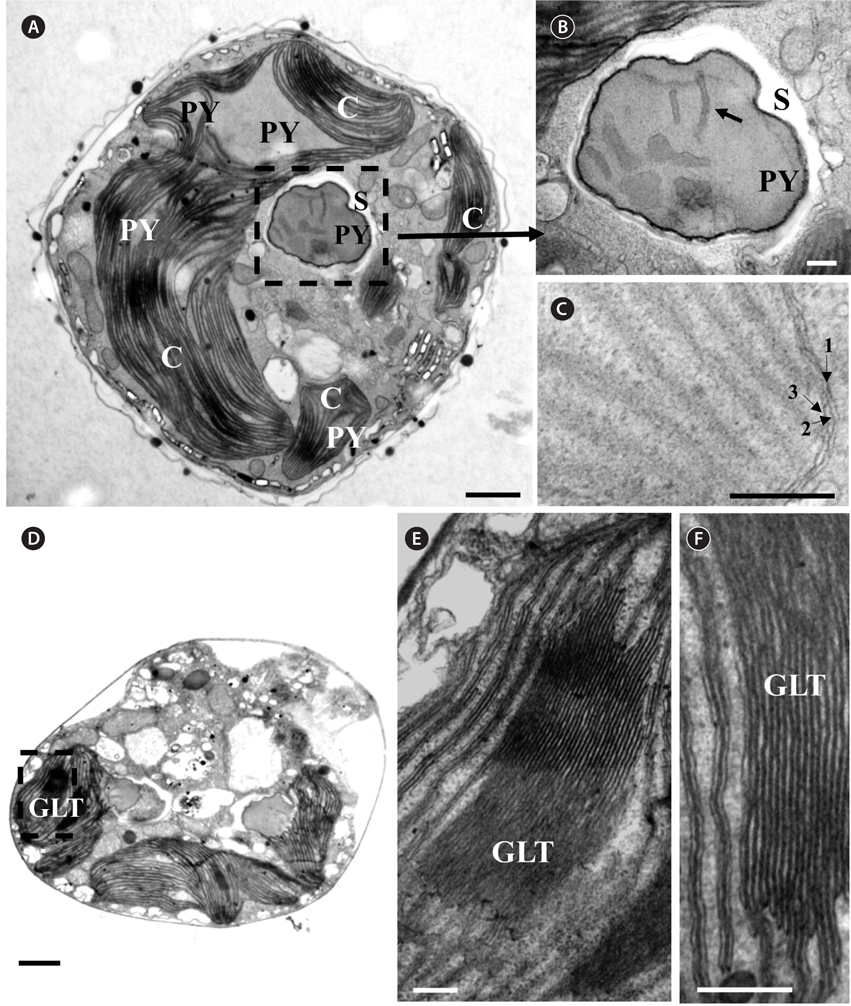
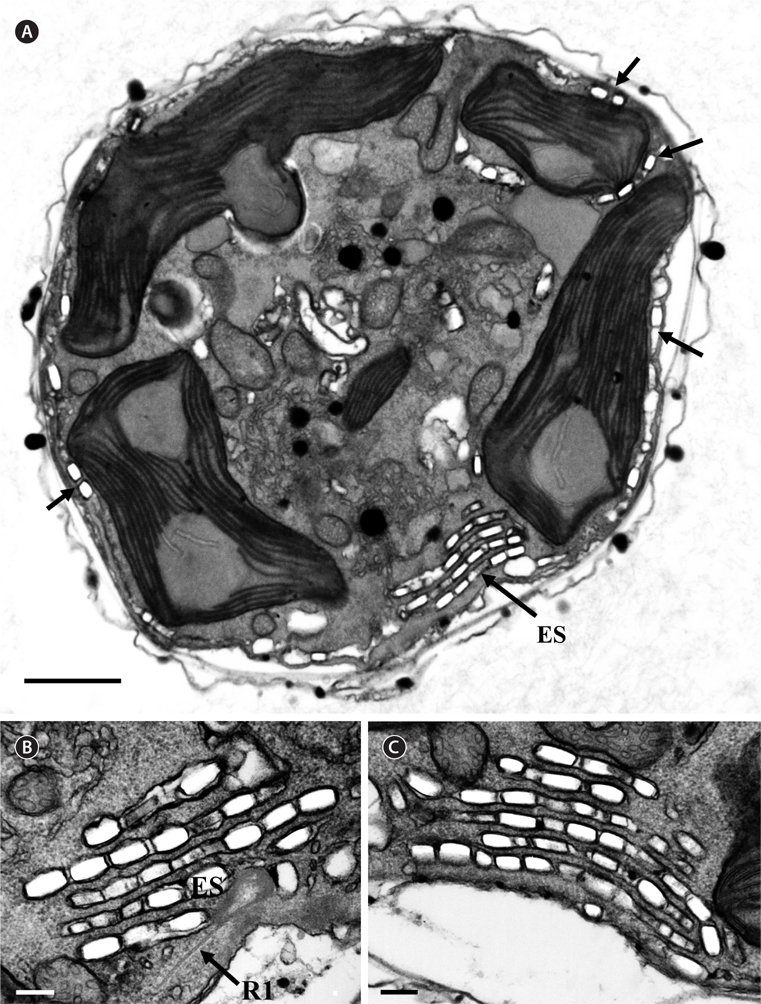
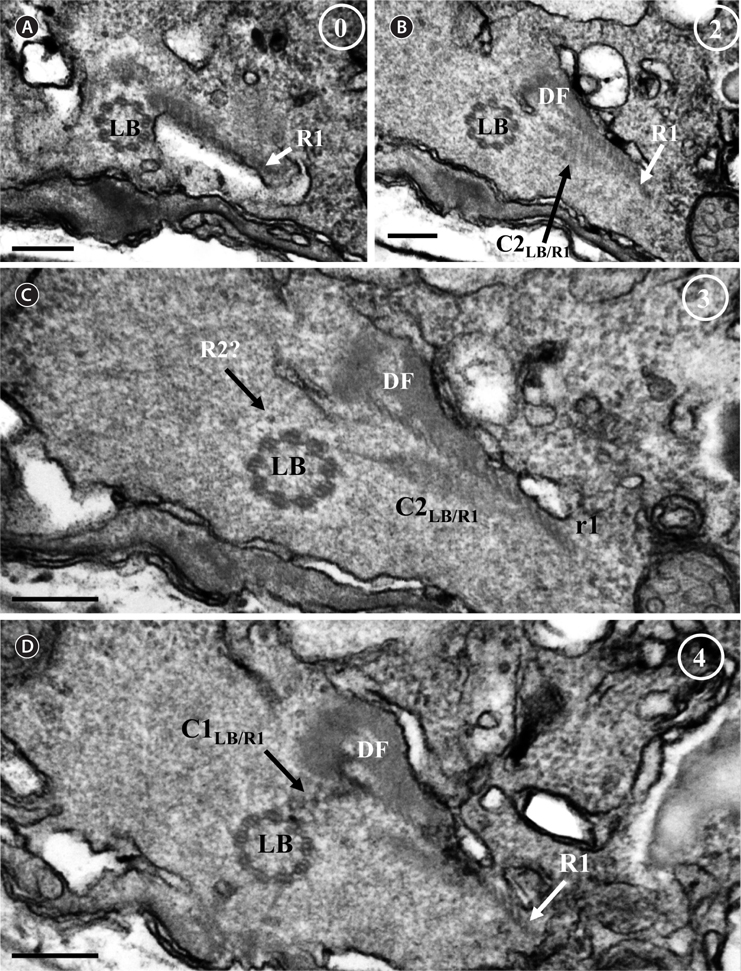
Thin TEM sections showed the main features of the cell, such as the chloroplasts, eyespot, fibrous vesicles, golgi body, mitochondria, nucleus, pyrenoid, and starch (Figs 5A-C & 6A). Longitudinal TEM serial sections showed the nucleus to contain many chromosomes, and its length and width were approximately half of the cell length (data not shown). The nucleus lacked nuclear chambers and a nuclear fibrous connective. Mitochondria were present in both central and peripheral parts (Fig. 5A-C). The golgi apparatus, composed of 6-10 stacked cisternae and fibrous vesicles, was located near the nucleus (Fig. 5A & B). Chloroplasts with complex lobes were located predominantly near the cell surface. These chloroplasts typically had three thylakoids per lamella (Fig. 6C & F), but arrangements of five, six, or more thylakoids per lamella (i.e., grana thylakoid associations) were also seen (Fig. 6D-F, Appendix 2B & C). All sectioned cells showed extensive “grana” formation (Fig. 6D-F, Appendix 2B & C). Three evenly spaced membranes enclosed each chloroplast (Fig. 6C). The chloroplasts formed a peripheral network that comprised 3-4 pyrenoids, each surrounded by a hemispherical starch grain (Fig. 6A & B, Appendix 2A). Underneath the sulcus, there was a large and conspicuous eyespot comprising approximately ~7 cisternae with brick-like material (Fig. 7A-C). The length and width of the crystalline bricks were 100-230 nm and 50-120 nm, respectively. The eyespot was located near the flagellar apparatus (Fig. 7B). A single row of microtubules (i.e., Root 1, R1) was observed in the narrow space between the outermost cisterna and the cell membrane of the sulcal region (Fig. 7B). The flagellar apparatus of several cells was examined in TEM serial sections (Figs 8-12), and a diagrammatic 3D-reconstruction was generated (Fig. 13). The transverse basal body (TB) and longitudinal basal body (LB) formed an angle of approximately 135˚ (Fig. 8A), as estimated from the serial sections. A striated collar surrounded the exit aperture of each flagellar canal (longitudinal striated collar and transverse striated collar [TSC]) (Figs 8 & 12). In the flagellar root system, a large multi-membered microtubular root 1 (R1, longitudinal microtubular root) was located to the left of the LB (Figs 8-10 & 12) and extended underneath the sulcus toward the antapex (not shown). A single-stranded microtubular root (R2) was associated obliquely with the right side of the LB (Figs 8D, 9C, 10C & D). The transverse microtubular root (R3) was attached to the right side of the TB (Fig. 10). The transverse microtubular root extension structure was located near the TB and R3 (Fig. 10C & D). The transverse striated root and the transverse striated root microtubule of root 4 were located near the TB (Figs 8E, 11 & 12A). The two basal bodies were connected by a small striated connective, the basal body connectives (Fig. 8D). A number of fibrous connectives or fibres were associated with the various roots. Thus, the R1 root was attached to the LB by two fibrillar connectives, the C1LB/R1 and C2LB/R1, respectively (Fig. 9). The C1LB/R1 was situated slightly posterior to C2LB/R1 and attached to the two rightmost microtubules of the R1 and one of the LB triplets. It was relatively short and only visible in two consecutive sections. The C2LB/R1 showed a distinct striation pattern and was intimately associated with the ventral surface of the R1 (Fig. 9C). The R1 and R4 roots were interlinked by a striated root connective (Fig. 8D & E). A dorsal fibre, with almost the same striation pattern as the striated fibre of R4, was located on the dorsal side of the R1 (Figs 10B, C, E & 12C). Transverse TEM serial sections showed the pusule system (PU) (Fig. 11). In the right-antapical side view, the PU was located next to the TBs and TSC (Fig. 11). We serially sectioned 5 whole cells transverally and longitudinally, but peduncle, lamellar body and rhizocysts were not observed.
[Fig. 6.] Micrographs of Ansanella granifera AGSW10 gen. et sp. nov. take by transmission electron microscopy. (A) The stalked pyrenoid (PY) (dashed box) is surrounded by a starch sheath (S). (B) Same figure enlarged, showing the stalked PY, S, and thylakoids (arrow). (C) The chloroplasts are bounded by three membranes (arrows); each lamella possesses three thylakoids. (D-F) Chloroplast lobe, showing the grana-like thylakoids (GLT, dashed box) and thylakoids mostly in triplets. C, chloroplasts. Scale bars represent: A & D, 1 μm; B, C, E & F, 0.2 μm.
[Fig. 8.] Ansanella granifera AGSW10 taken by transmission electron microscopy. (A) Longitudinal section of the cell showing the basal bodies (longitudinal basal body [LB], transverse basal body [TB]) and the eyespot (ES). (B-E) Flagellar apparatus. Non-adjacent, nearly longitudinal serial sections proceeding from left to right. The encircled numbers are section numbers. Micrograph showing relative positions of the LB, TB, Root 1 (R1), putative Root 2 (R2), Root 4 (R4), striated root connective (SRC), basal body connectives (bbc), and longitudinal striated collar (LSC). Scale bars represent: A-E, 0.2 μm.
[Fig. 13.] Diagrammatic reconstruction of the flagellar apparatus of Ansanella granifera AGSW10 gen. et sp. nov., based mainly on transverse serial sections (70-nm-thick sections). LB, longitudinal basal body; TB, transverse basal body; LSC, longitudinal striated collar; TSC, transverse striated collar; R1, root 1, longitudinal microtubular root; R2, root 2, single-stranded microtubular root; R3, root 3, transverse microtubular root; R4, root 4, transverse striated root (TSR) + transverse striated root microtubule (TSRM); SRC, striated root connective; C1LB/R1, connective 1 linking LB and R1; C2LB/R1, connective 2 linking LB and R1; bbc, basal body connective; TMRE, transverse microtubular root extension; DF, dorsal fiber.
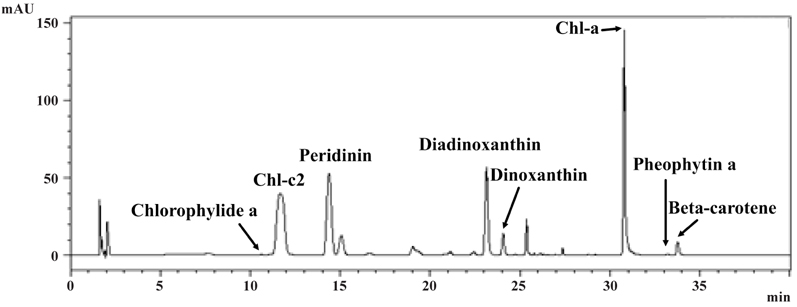
Pigment analysis of A. granifera AGSW10 showed chlorophyll a as the major pigment, and chlorophyllide a, chlorophyll c2, peridinin, diadinoxanthin, dinoxanthin, pheophytin a, and beta-carotene as accessory pigments (Fig. 14).
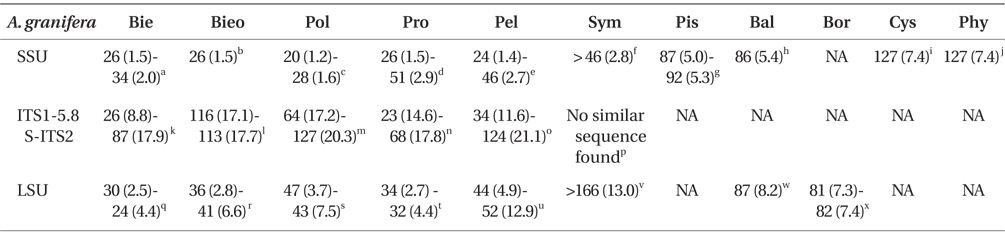
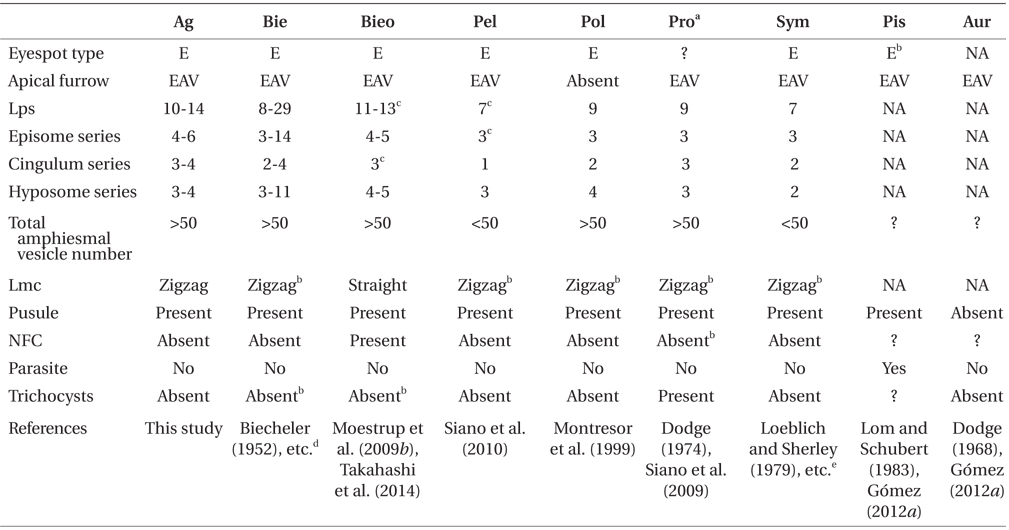
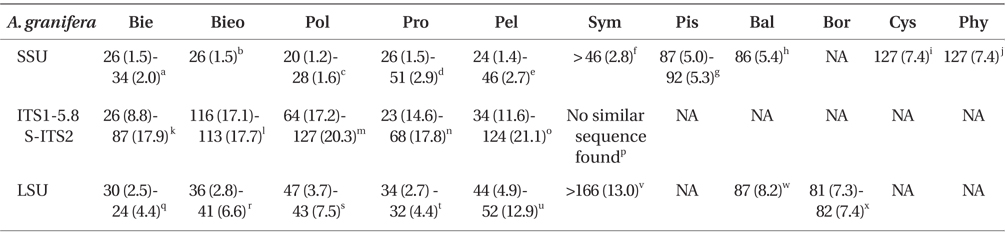
DNA sequences of A. granifera gen. et sp. nov. The SSU, ITS1, 5.8S, ITS2, and D1-D3 LSU rDNA sequences of A. granifera AGSW10 (GenBank accession Nos. HG529978-HG529980, 3,663-bp product) were only 1 base pair different from those of A. granifera AGSW12 (HG792066, 3,663-bp product). When properly aligned, the SSU rDNA sequence of A. granifera differed by 1.2-7.4% from those of the other genera included in the order Suessiales (i.e., Biecheleria, Biecheleriopsis, Polarella, Protodinium [it should be noted that strains such as CCMP 419 and CCMP 420 appear to belong to Biecheleriopsis adriatica, not to Protodinium], Pelagodinium, Symbiodinium, Piscinoodinium, Baldinia, Cystodinium, and Phytodinium). In addition, the ITS1, 5.8S, and ITS2 rDNA sequences of A. granifera differed by >8.8% from those of the other genera included in the family Suessiaceae (i.e., Biecheleria, Biecheleriopsis, Polarella, Protodinium, and Pelagodinium). A. granifera had a 51-base pair fragment in domain D2 of the large subunit of ribosomal DNA, which has not been found in genus Biecheleria. The D1-D3 LSU rDNA sequences of A. granifera differed by >2.5% from those of other genera included in the order Suessiales (i.e., Biecheleria, Biecheleriopsis, Polarella, Protodinium, Pelagodinium, Symbiodinium, Baldinia, and Borghiella) (Table 1).
When properly aligned, the SSU rDNA sequence of A. granifera differed by 1-2% from those of the 3 most closely related species-Polarella glacialis (EF417317), Pelagodinium bei (U37406) (to be spelled with a single i, commemorating A.W.H.Bé), and Protodinium simplex (U41086). The ITS1-5.8S-ITS2 rDNA sequence differed by 16-19% from those of the 3 most closely related species-Gymnodinium corii (AF318226), Protodinium simplex (AY686651), and Pelagodinium bei (DQ195358). Furthermore, the D1-D3 LSU rDNA sequence differed by 3-4% from those of the 3 most closely related species-“Protodinium sp.” CCMP419 (EF205015), Biecheleriopsis adriatica (EU857537), and Gymnodinium sp. CCMP425 (EF205006).
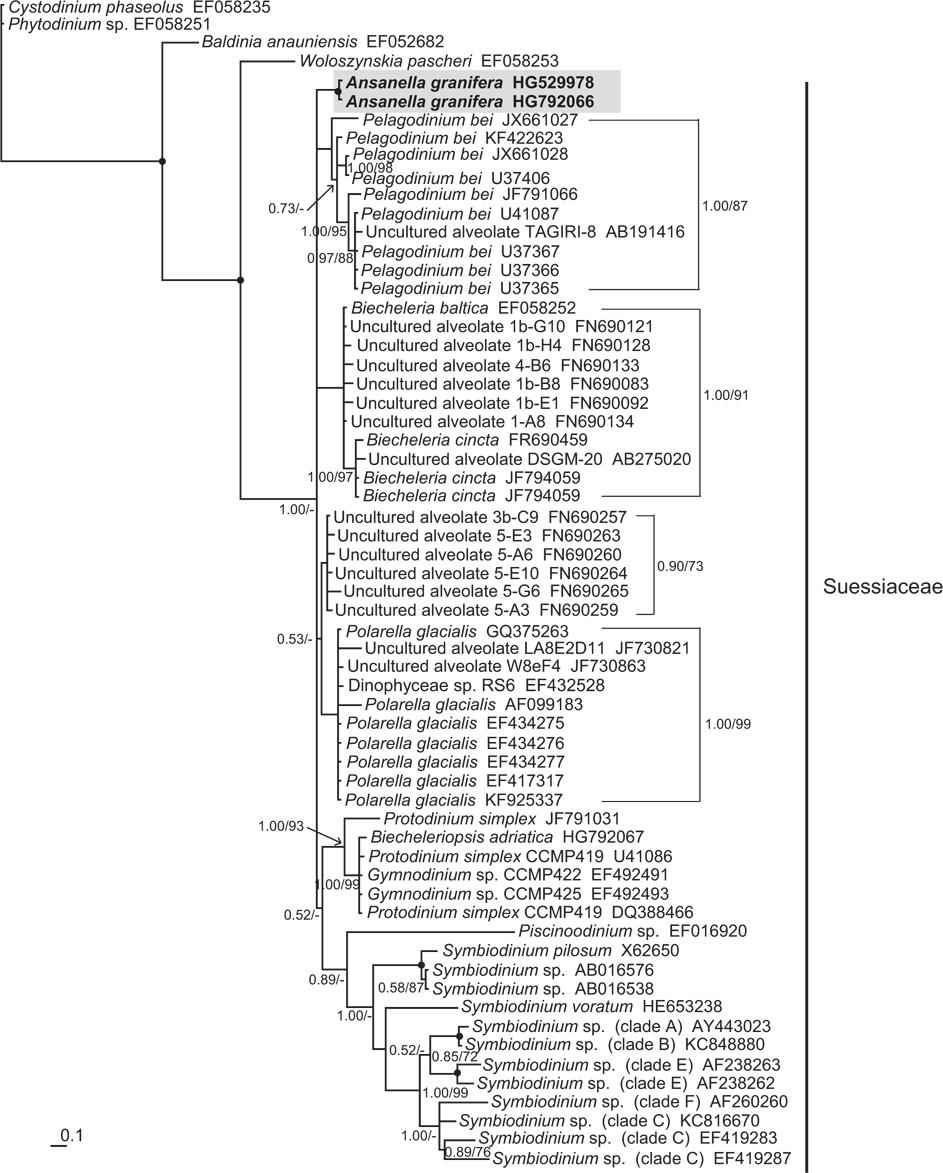
In the phylogenetic trees based on the SSU and LSU rDNA sequences of dinoflagellates, A. granifera belonged to the large clade of the family Suessiaceae (Figs 15 & 16). In the tree based on SSU rDNA sequences, A. granifera formed an independent clade with other genera in the family Suessiaceae (Fig. 15). In the tree based on D1-D3 LSU rDNA sequences, A. granifera also formed a subclade with Symbiodinium spp. (Fig. 16). This relationship was supported in terms of posterior probability (66%). However, it was not significantly supported in terms of ML bootstrap values. Also, the clade containing A. granifera strains was clearly divergent from the clades containing the other genera in the family Suessiaceae (Fig. 16).
The newly isolated cells moved in straight lines or with a helicoidal mode. They sometimes stopped suddenly, and then resumed movement very quickly, with a jumplike action. They grew at salinities > 10 or <35, but died in fresh water. Thus, this dinoflagellate is an estuarine and marine species.
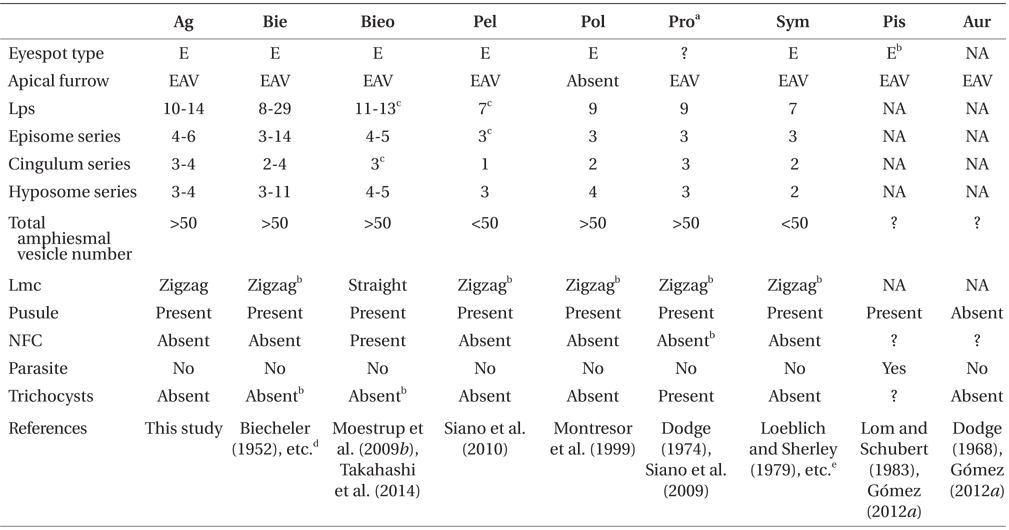
Ansanella granifera gen. et sp. nov. belongs to the family Suessiaceae of the order Suessiales; it has 10-14 latitudinal series of AVs, thus meeting the criteria for inclusion in the order Suessiales (Fensome et al. 1993, Kremp et al. 2005). In addition, A. granifera has eyespot type E (i.e., a series of cisternae, with brick-like contents) and a single elongated apical vesicle, which are key characters of the family Suessiaceae (Moestrup and Daugbjerg 2007, Moestrup et al. 2009a), whereas members of the family Borghiellaceae, with the two genera Baldinia and Borghiella have eyespot of type B (i.e., eyespots containing carotenoid globules located within the chloroplast) (Hansen et al. 2007, Moestrup and Daugbjerg 2007, Moestrup et al. 2008, 2009a).
The morphology of Ansanella gen. nov. is different from that of other genera in or possibly related to the family Suessiaceae (i.e., Biecheleriopsis, Pelagodinium, Polarella, Protodinium, Symbiodinium, perhaps Aureodinium and Piscinoodinium) (Lohmann 1908, Freudenthal 1962, Dodge 1967, Lom 1981, Montresor et al. 1999, Moestrup et al. 2009a, 2009b, Siano et al. 2010, Gómez 2012a): Cells of Ansanella gen. nov. possess a pusule, while a pusule has not been reported in Aureodinium (Dodge 1967); Ansanella lacks a nuclear fibrous connective, which is present in Biecheleriopsis (Moestrup et al. 2009b). Further, B. adriatica and A. granifera showed other morphological difference in the shape of margin in cingulum. In A. granifera, the cingular margin formed a zigzag line, while in B. adriatica it forms a straight line, especially on the dorsal side; Ansanella has a single elongated apical vesicle, which is absent in Polarella (Montresor et al. 1999); Ansanella has a larger number of latitudinal plate series (10-14) than Pelagodinium, Polarella, Protodinium, and Symbiodinium (7-9); Ansanella also has a larger total number of AVs (>50) than Pelagodinium, Symbiodinium (<50); Ansanella lacks trichocysts, which are present in Protodinium (Dodge 1974, however, the identity of the organism illustrated in the paper needs to be confirmed); Ansanella is a free-living form, and lacks rhizocysts, while Piscinoodinium is parasitic with rhizocysts (Lom and Schubert 1983); the morphology of Ansanella is similar to that of Biecheleria (Moestrup et al. 2009b). However, Ansanella has a 51-base pair fragment in domain D2 of the LSU rDNA, which is absent in the genus Biecheleria (Moestrup et al. 2009b, Takahashi et al. 2014).
A. granifera has grana-like thylakoids. In the Suessiales sensu lato, these have only been reported previously in the parasite Piscinoodinium (Dodge 1968, Lom and Schubert 1983), whereas grana-like thylakoids are present in other orders, including Gonyaulacales (Alexandrium pseudogonyaulax, Pyramidodinium atrofuscum, Pyrocystis lunula, Gonyaulax spinifera) and Thoracosphaerales (Theleodinium calcisporum) (Dodge 1975, Hansen et al. 1996, Horiguchi and Sukigara 2005, Craveiro et al. 2013).
In the family Suessiaceae, the Type E eyespot has been reported previously in Biecheleriopsis adriatica Moestrup, Lindberg and Daugbjerg; Symbiodinium natans Hansen and Daugbjerg; Symbiodinium voratum Jeong, Lee, Kang and LaJeunesse; Polarella glacialis Montresor, Procaccini and Stoecker; Pelagodinium bei Siano, Montresor, Probert and de Vargas; Biecheleria pseudopalustris Moestrup, Lindberg and Daugbjerg; Biecheleria baltica Moestrup, Lindberg and Daugbjerg; Biecheleria natalensis (Horiguchi and Pienaar) Moestrup; Biecheleria cincta (Siano, Montresor and Zingone) Siano; and Biecheleria brevisulcata Takahashi & Iwataki (Table 2). The eyespot of A. granifera consists of 7 layers of cisternae, identical or similar to those of B. brevisulcata (4), P. glacialis (5), P. bei (6), B. adriatica (6), S. natans (6), S. voratum (6), B. natalensis (6), Prosoaulax lacustris (6), B. pseudopalustris (7), B. cincta (7), and B. baltica (9) (Horiguchi and Pienaar 1994b, Calado et al. 1998, Montresor et al. 1999, Kremp et al. 2005, Hansen and Daugbjerg 2009, Moestrup et al. 2009a, 2009b, Siano et al. 2010, Fig 15 in Kang et al. 2011a, Jeong et al. 2014, Takahashi et al. 2014). Whether Type E eyespot occurs in other species of the family Suessiaceae requires further investigation. It is a very unusual type of eyespot, and it has been suggested that the crystalline layers may function as a quarter-wave receptor (Foster and Smyth 1980, Horiguchi and Pienaar 1994b). Thus, the function of the eyespot in A. granifera merits further investigation. The actual photoreceptor has never been identified in any dinoflagellate.
The flagellar apparatus of Ansanella consists of typical dinoflagellate components, that is, R1-R4 flagellar roots and fibrous collars around the flagellar canals. It is basically similar to that described for Symbiodinium natans Hansen and Daugbjerg, with respect to the various connectives interlinking the flagellar roots and the basal bodies, with one notable difference, the presence of peduncle microtubules near the basal bodies in Symbiodinium natans (Hansen and Daugbjerg 2009). Also, the flagellar apparatus of A. granifera differs from S. natans by the presence of a ventral fiber. The possible presence of a peduncle and ventral fiber in the other species in the family Suessiaceae needs to be determined.
The SSU, ITS1, 5.8S, ITS2, and D1-D3 LSU rDNA sequences of the genus Ansanella clearly differ from those of any other genus in the family Suessiaceae. In the SSU and LSU phylogenetic trees, A. granifera belongs to the family Suessiaceae. However, in the LSU phylogenetic tree, the two strains of A. granifera formed a clade that is divergent from clades containing other genera in the family Suessiaceae. This clear divergence shows concordance between morphological and genetic characteristics. Therefore, on the basis of the morphological and molecular data, we propose that A. granifera to be a new species within a new genus.
There are indications that the order Suessiales may comprise not only the fish parasite Piscinoodinium but also species of the order Phytodiniales, members of which are coccoid freshwater dinoflagellates. Thus Logares et al. (2007) found both Cystodinium phaseolus and Phytodinium sp. of the Phytodiniales to be phylogenetically related to the Suessiales. Studies are underway to examine the ultrastructure of Cystodinium to determine whether Phytodiniales should be included in the Suessiales or retained as a separate order (Moestrup, unpublished). If the former turns out to be the case, the International Code of Nomenclature states that the priority rule is not mandatory for names above the level of family and the most suitable order name can therefore be applied (McNeill et al. 2012, Art. 16.4. Note 2).








![Ansanella granifera AGSW10 taken by transmission electron microscopy. (A) Longitudinal section of the cell showing the basal bodies (longitudinal basal body [LB], transverse basal body [TB]) and the eyespot (ES). (B-E) Flagellar apparatus. Non-adjacent, nearly longitudinal serial sections proceeding from left to right. The encircled numbers are section numbers. Micrograph showing relative positions of the LB, TB, Root 1 (R1), putative Root 2 (R2), Root 4 (R4), striated root connective (SRC), basal body connectives (bbc), and longitudinal striated collar (LSC). Scale bars represent: A-E, 0.2 μm.](http://oak.go.kr/repository/journal/13610/JORHBK_2014_v29n2_75_f008.jpg)
![Ansanella granifera AGSW10 taken by transmission electron microscopy. Nonadjacent longitudinal serial sections of the flagellar apparatus. The cell is seen from the outside, and the sectioning proceeds from right to left. The encircled numbers are section numbers. (A-D) Micrograph showing relative positions of the longitudinal basal body (LB), transverse basal body (TB), Root 1 (R1), putative Root 2 (R2), Root 3 (R3), Root 4 (R4), dorsal fiber (DF), and microtubular extension (transverse microtubular root extension [TMRE]). The striation pattern of the DF is very distinct. Notice also flagellar root R3 and its microtubular extension (TMRE). (E) Enlargement from Fig. 10C. Scale bars represent: A-D, 0.2 μm.](http://oak.go.kr/repository/journal/13610/JORHBK_2014_v29n2_75_f010.jpg)
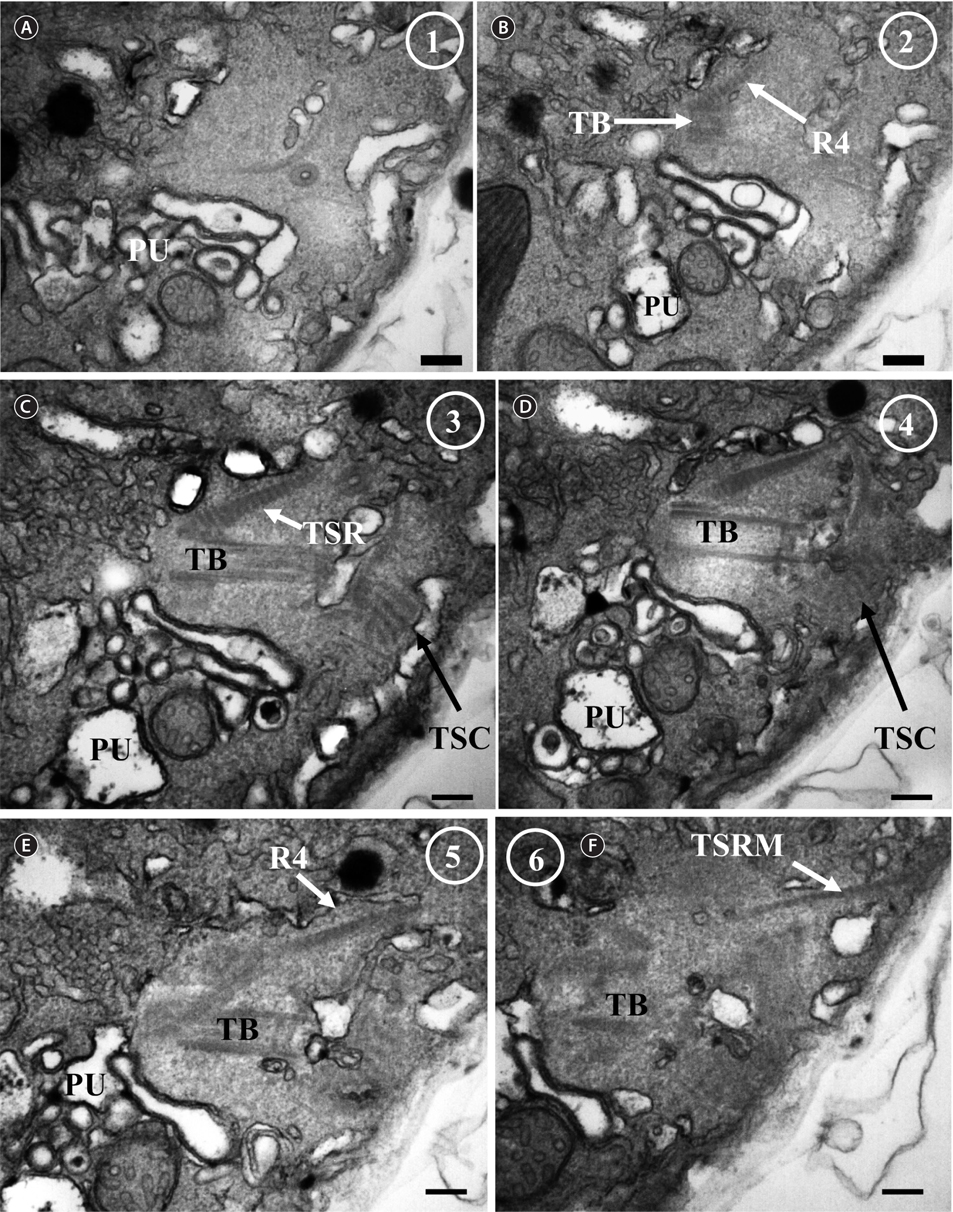
![Ansanella granifera AGSW10 taken by transmission electron microscopy. Nonadjacent serial sections. The sulcus region in transverse section, the sectioning moves from posterior to anterior. The encircled numbers are section numbers. (A-D) Micrograph showing relative positions of the longitudinal basal body (LB), transverse basal body (TB), Root 1 (R1), Root 4 (R4, transverse striated root [TSR] + transverse striated root microtubule [TSRM]), dorsal fiber (DF), transverse striated collar (TSC), and eyespot (ES). The R1 flagellar root is located in the narrow space between the eyespot and the cell surface. Scale bars represent: A-D, 0.2 μm.](http://oak.go.kr/repository/journal/13610/JORHBK_2014_v29n2_75_f012.jpg)