Many red algal morphospecies comprise a grouping of several genetic entities, which are usually referred to as cryptic species (Le Gall and Saunders 2010). Our ability to distinguish these genetic entities using traditional morphological diagnoses, while commendable, may not be achievable (Verbruggen 2014). This is especially true for the more simply constructed red algae (e.g., Zuccarello et al. 2011).
The ubiquity and taxonomy of the ‘simple’ red algae once considered in the class Bangiophyceae is slowly being resolved (e.g., Zuccarello et al. 2008, 2010, 2011, Necchi et al. 2013). The old Bangiophyceae is now subdivided into six separate classes (Yoon et al. 2006), one of which is the Stylonematophyceae. The Stylonematophyceae is comprised of 14 microscopic genera that are mostly marine but two are freshwater and one is terrestrial (Zuccarello et al. 2008). Most of the genera are filamentous and two are unicellular (Rhodosorus and Rhodospora). Stylonema alsidii is probably the most ubiquitous species in the class (Zuccarello et al. 2008). New genera (Purpureofilum and Rhodaphanes) have also been described from recent collections (West et al. 2005, 2007).
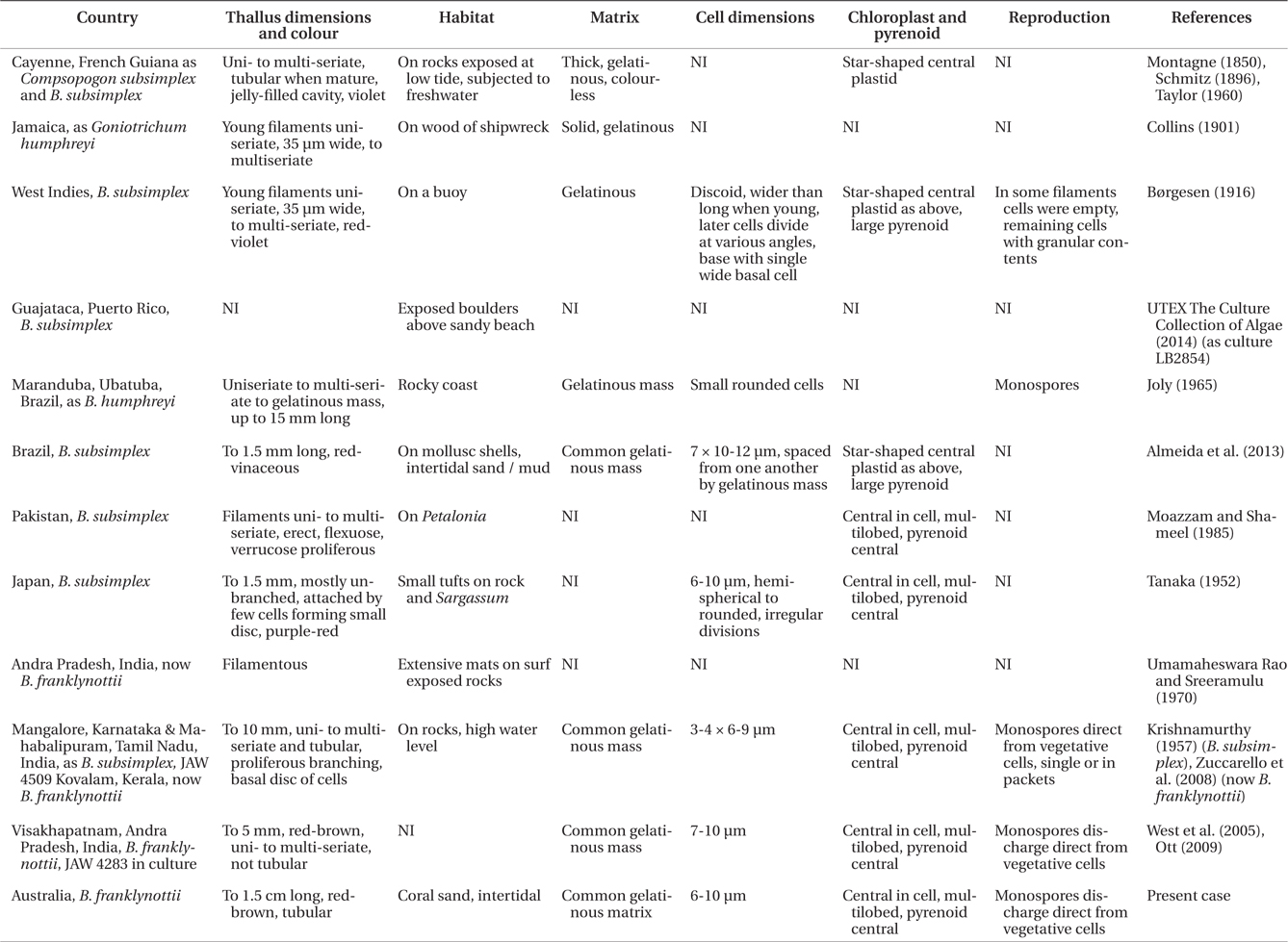
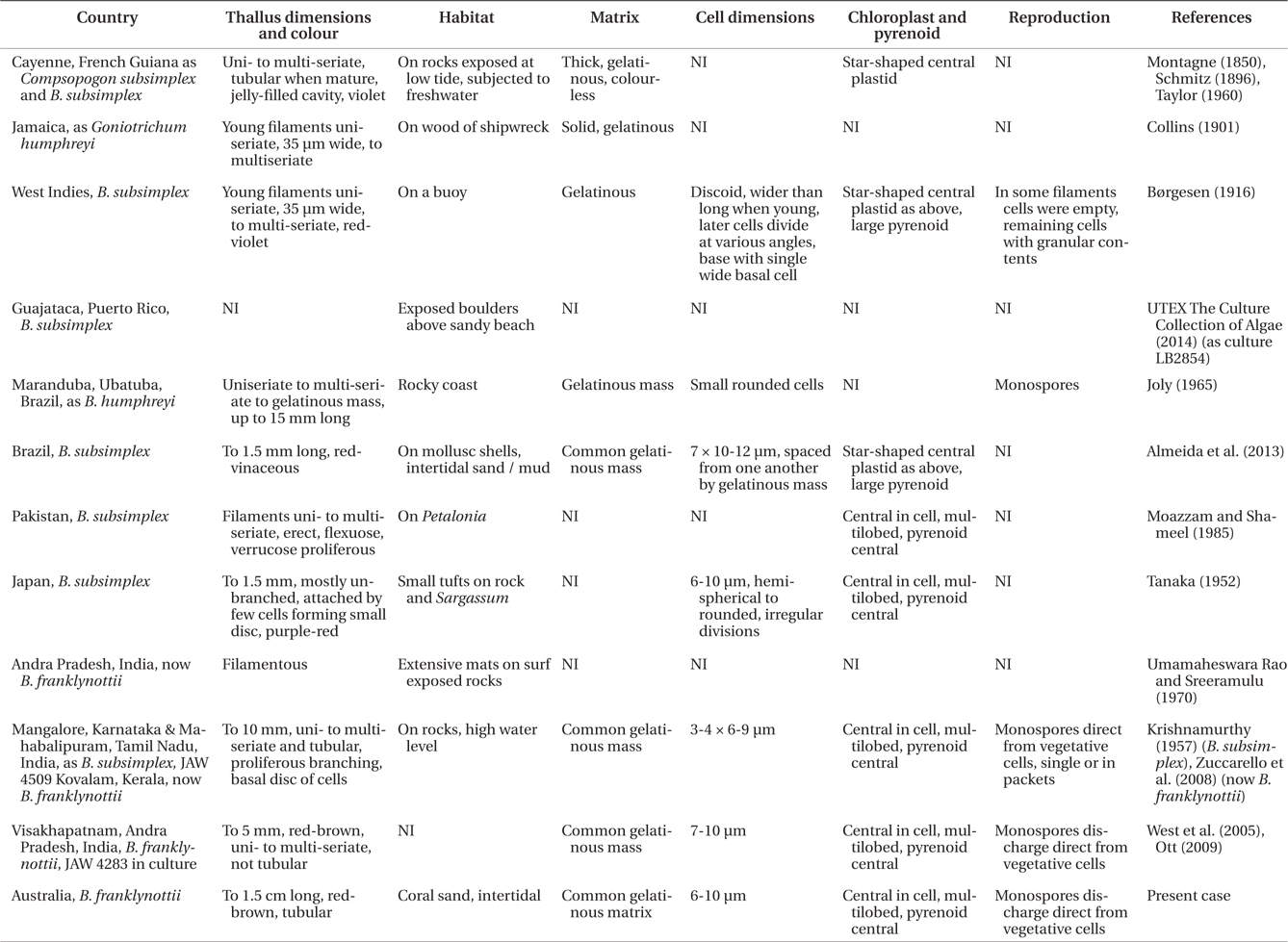
Bangiopsis is a branched filamentous genus up to 1 cm long. The genus has a complicated taxonomic and nomenclatural history (Krishnamurthy 1957, Silva et al. 1996, Guiry and Guiry 2014). Bangiopsis subsimplex (Montagne) F. Schmitz is the type species originally described by Montagne (1850) as Compsopogon subsimplex Montagne based on specimens collected by Leprieur in Cayenne, French Guiana. It was transferred to Bangiopsis by Schmitz (1896). Borgesen (1916) reported a similar specimen from Christianssted, St. Croix, Danish West Indies that he designated as B. subsimplex, however, Collins and Hervey (1917) thought it to be more like Goniotrichum humphreyi Collins found growing on wood in St. Annes Bay, Jamaica (Collins 1901). Hamel (1929) transferred that to Bangiopsis humphreyi (Collins) G. Hamel but this is now treated as a synonym of Bangiopsis dumontioides (P. L. Crouan & H. M. Crouan) V. Krishnamurthy (Krishnamurthy 1957). Bangiopsis subsimplex was also collected in Puerto Rico at Guajataca Beach by Rintoul and Sherwood in 1997 providing the first specimens of the genus used for molecular analysis (Muller et al. 2001). In 2005 it was also collected by David Ballantine from the same site, cultured by Franklyn Ott and placed in the UTEX The Culture Collection of Algae as LB 2854.
Joly (1965) recorded B. subsimplex in São Paulo, Brazil. This species was also reported by Almeida et al. (2013) using cultured samples from Baía la de Todos os Santos, Bahia, Brazil comparing it with several similar genera of the Stylonematales. Brasileiro et al. (2009) listed, without explanation, B. dumontioides from Cabo Frio, Brazil.
Krishnamurthy (1957) obtained B. subsimplex samples from Mangalore, Karnataka and near Chennai, Tamil Nadu, India. Umamaheswara Rao and Sreeramulu (1970) found B. subsimplex at Visakhapatnam, Andra Pradesh, India. Although this has not been further verified, Krishnamurthy (1957) stated that a Natural History Museum (London) specimen of Bangia fergusoni Grunow from Sri Lanka is identical to B. subsimplex. Moazzam and Shameel (1985) observed B. subsimplex on Petalonia sp. in Pakistan. Tanaka (1952) recorded B. subsimplex growing on rocks and Sargassum sp. near Satoura, Japan. Kapraun and Bowden (1978) recorded B. humphreyi in Fiji but South and Skelton (2003) referred to it as B. subsimplex.
The basic problem with species and generic identification in the Stylonematophyceae is that they are not easily discernible from nomenclatural descriptions, and without molecular data are even more uncertain. Taxonomic progress requires careful field collections, culture and molecular observations. Even though Bangiopsis records are widespread it is difficult to infer the morphological variation from previous reports. Many of the description lack key anatomical details and this makes identification of species and understanding species distribution impossible.
We collected an isolate of Bangiopsis in Australia. Molecular analysis of this isolate compared to available isolates from other parts of the world indicate that it belongs to a new species.
Pale brown / red microscopic filamentous specimens were collected by Richard Wetherbee in a shallow sandy tidal pool on Jul 6, 2013 just north of Noah Beach car park (ca. 16˚07.896′ S, 145˚27.302′ E), Cape Tribulation, Queensland, Australia. Initially sand samples were placed in culture flasks with seawater from the site and held at about 20 ± 2℃ on a rotary shaker at approximately 25 rpm and 30 ℃mol photons m-2 s-1 cool white LED lighting at 10 : 14 LD daily cycle. Multiseriate filaments were checked for epiphytes and isolated by excising short segments about 1 mm long. These were placed in 50 × 70 mm culture dishes with 1/4 strength Modified Provasoli’s Medium (20 mL enrichment per litre sterile seawater) to supress epiphyte growth. GeO2 and sodium penicillin G were added to inhibit diatoms and cyanobacteria, respectively (West 2005). For slower growth light was reduced to about 6-7 µmol photons m-2 s-1. Other conditions were maintained as stated above. The culture was designated as JAW 4835.
Molecular analyses were done on the Australian specimen and two other isolates: UTEX LB 2854 from Guajataca, Puerto Rico (Ott MO 7041) and Ott MO 7045 (isolated Nov 27, 2004 from specimens forming distinct bands on rocks from Fisherman’s Cove, Chennai, India and obtained by V. Krishnamurthy, JAW 4567).
Total DNA was isolated from silica gel-dried cultured material using a modified CTAB procedure (Zuccarello and Lokhorst 2005). Amplification and sequencing of the plastid-encoded large subunit of the ribulose bisphosphate carboxylase / oxygenase gene (rbcL) used amplification primers presented in Freshwater and Rueness (1994). The psbA (photosystem II reaction center protein D1) gene was amplified using the primers described in Yoon et al. (2002). Amplified products were checked for correct length, purity and yield on 1% agarose gels. Polymerase chain reaction products were cleaned using ExoSAP-IT (USB, Cleveland, OH, USA) and commercially sequenced (Macrogen Inc., Daejeon, Korea). A selection of members of the Stylonematophyceae and Compsopogonophyceae were used from previous publications (Zuccarello et al. 2008, 2011). Outgroups used in all analyses were Porphyridium aerugineum Geitler and Flintiella sanguinaria Ott, members of the Porphyridiophyceae and Compsopogon caeruleus (Balbis ex C. Agardh) Montagne of the Compsopogonophyceae.
Sequences were edited, assembled and aligned using the Geneious software package (Biomatters, available from http://www.geneious.com/). Alignment was straight forward as no gaps were found in the data set and the two genes were combined. The program Modeltest version 3.7 (Posada and Crandall 1998) was used to find the model of sequence evolution that best fit the data set by an Akaike Information Criterion (AIC) (Posada and Crandall 2001). Maximum likelihood (ML) was performed with RAxML 7.2.8 (Stamatakis 2006). RAxML was performed with all threes codons partitioned and the GTR + gamma model and 500 non-parametric bootstrap replicates (Felsenstein 1985).
Bayesian inference was performed with MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003). Analyses consisted of two independent simultaneous runs of one cold and three incrementally heated chains, and 3 × 106 generations with sampling every 1,000 generations. Codons were partitioned. The log files of the runs were checked with Tracer v1.5 (Rambaut and Drummond 2007) and a burn-in sample of 100 trees was removed from each run before calculating the majority rule consensus tree.
A field specimen grown in culture showed a uniseriate filament branching and forming multiseriate upper sectors that became tubular with a single peripheral layer of cells (Fig. 1A & C). Thalli were up to 1.5 cm long and red coloured (Fig. 1A). Cell divisions in the uniseriate section were initially transverse and then longitudinal to irregular in orientation forming biseriate and multiseriate sectors (Fig. 1B). Older tubular branches were composed of a single peripheral layer of cells containing multicellular packets of a polyhedral appearance with a thick colorless matrix (Fig. 1C) and some of these vegetative cells were released directly as spores (Fig. 1B). Each cell had a single nucleus laterally displaced by a single multilobed plastid with a large central pyrenoid (Fig. 1C). In this Bangiopsis culture small dark brown deposits (10-15 ℃m in diameter) with a lighter brown central area were common (Fig. 1C, arrowheads). These were similar to brown deposits that were composed of manganese and other elements, as observed previously in cultures of other algae (West et al. 2013).
The spherical spores were 9-10 ℃m in diameter and displayed slow rotation combined with gliding movement before settling on a substrate. No amoeboid movement was seen. Germination was initiated by a series of cell divisions, with second divisions usually at right angles to the first divisions and then dividing in various planes to form a multicellular aggregate enclosed in a thin matrix (Fig. 1D). Erect shoots normally developed from aggregates of 12 or more cells (Fig. 1D, two arrowheads) but sometimes an erect filament formed directly from an undivided spore (Fig. 1D, single arrowhead). In low light (6-7 ℃mol photons m-2 s-1) the basal system was extensive and erect filaments formed late (Fig. 1D). In brighter light (20-22 ℃mol photons m-2 s-1) erect branching from the basal system was enhanced (Fig. 1E).
Isolate MO1560 (JAW 4283) was analysed for low molecular weight carbohydrates (sorbitol 1,102 mmol kg-1 DW and digeneaside 20 mmol kg-1 DW, floridoside was not detected). Isolate MO70450 (JAW 4509) had sorbitol 485 mmol kg-1 DW, digeneaside 14 mmol kg-1 DW and floridoside 2 mmol kg-1 DW (Ulf Karsten, personal communication).
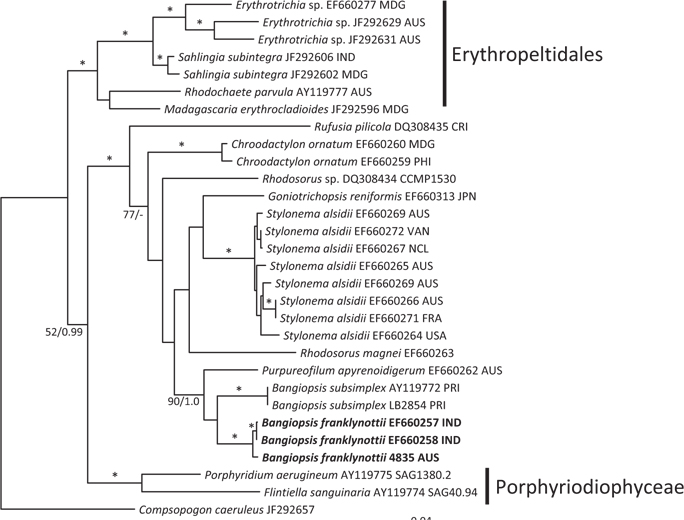
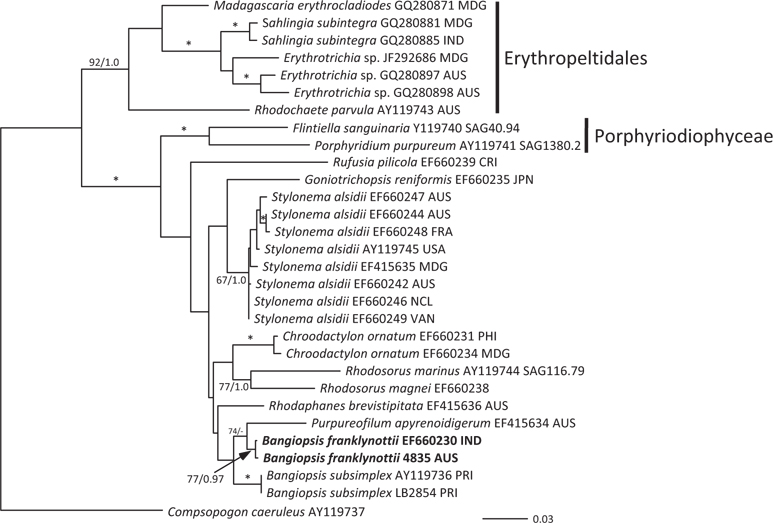
The topologies of the two gene trees are similar to previous phylogenies of the class Stylonematophyceae, and also similar between the ML and Bayesian analysis. The class Porphyridiophyceae and order Erythropeltidales are both well supported, and Rufusia pilicola is sister to the remaining Stylonematophyceae. The two different plastid gene phylogenies clearly show the Australian isolate groups with Bangiopsis isolates from India (Figs 2 & 3) and are separate from the Puerto Rico samples. The genus Bangiopsis while not supported within these datasets appears to be closely related to Purpureofilum apyrenoidigerum. The well supported differences between the isolates from the Indian and Pacific Oceans versus the samples from the Atlantic Ocean warrant the description of a new species for the non-Atlantic isolates.
[Fig. 2.] Maximum-likelihood (ML) topology (-log Ln = -9142.548) of rbcL gene data of select Stylonematophyceae, Porphyridiophyceae and Compsopogonophyceae. Composopogon caeruleus was used as an outgroup. * ≥95% ML bootstrap values and ≥0.95 Bayesian posterior probabilities. Other values associated with branches = RaxML bootstrap percentage/Bayesian posterior probabilities. Abbreviations following the species names and culture numbers: MDG, Madagascar; AUS, Australia; IND, India; CRI, Costa Rica; PHI, Philippines; JPN, Japan; VAN, Vanuatu; NCL, New Caledonia; FRA, France; USA, United States; PRI, Puerto Rico. Scale bar represents substitutions/site.
[Fig. 3.] Maximum-likelihood (ML) topology (-log Ln = -4505.523) of psbA gene data of select Stylonematophyceae. Composopogon caeruleus was used as an outgroup. * ≥95% ML bootstrap values and ≥0.95 Bayesian posterior probabilities. Other values associated with branches = RaxML bootstrap percentage / Bayesian posterior probabilities. Abbreviations following the species names and culture numbers: MDG, Madagascar; IND, India; AUS, Australia; CRI, Costa Rica; JPN, Japan; FRA, France; USA, United States; NCL, New Caledonia; VAN, Vanuatu; PHI, Philippines; PRI, Puerto Rico. Scale bar represents substitutions/site.
Pairwise differences between the two species also support this species distinction. Inter-species pairwise differences are over six times greater than the intra-species variation. For rbcL, intra-species variation is between 0.0-1.2%, while inter-species variation is 8.07-8.28%. For psbA intra-species variation is between 0.0-0.26% and interspecies variation 3.06%.
Description. Cells with single nucleus and a single central multilobed plastid containing a central pyrenoid. Basal attachment of thallus by a small multicellular disc giving rise to uniseriate and then multiseriate erect shoots by intercalary cell divisions. Mature thallus often becoming a hollow cylinder with a clear thick mucilaginous matrix in which cells are solitary or in packets. Monospores are discharged directly from vegetative cells along the axis.
Culture 4835 (CCMP 3416) was deposited in the National Center for Marine Algae and Microbiota (NCMA, https://ncma.bigelow.org/), 60 Bigelow Drive, PO Box 380, East Boothbay, Maine 04544, USA. A herbarium specimen was deposited at the National Herbarium of Victoria, Royal Botanic Gardens, Birdwood Ave, South Yarra VIC 3141, Australia (MEL 2371924). Genbank accession Nos: rbcL, KJ778645; psbA, KJ778647.
Our data clearly show that the isolates from Australia and the isolates from India are different from the Atlantic Ocean isolates from Puerto Rico. These genetic analyses clearly show that these isolates are distinct and probably have a common ancestor a long time ago. Based on rbcL mutation rate calibration from the red alga Caloglossa (Kamiya et al. 2004) the time to the most recent common ancestor for these two Bangiopsis species was approximately 69 (75-65) million years ago. Although this calibration from the Ceramiales must be taken with great caution, a great deal of time has passed since these species shared a common ancestor. As the type locality is Cayenne, French Guiana (Montagne 1850) in the Atlantic Ocean and these isolates are clearly distinct, we recognize the Indian and Australian isolates as a new species, B. franklynottii. This new species is not morphologically different from description of Bangiopsis subsimplex in the literature. There are probably several reasons for this. The first is that many descriptions of these small red algae are cursory and lack detail and certainly lack details that could only be revealed in culture (Table 1). Most of the characters overlap, for example cell dimensions, or are based on few specimens in which phenotypic plasticity was not investigated. Secondly, it is likely that morphology has not changed in this time. This lack of morphological characters to distinguish species, genera and even orders is becoming increasingly apparent with the influx of molecular data (West et al. 2008, Sutherland et al. 2011). It could be that many species can only be identified by using molecular data (Verbruggen 2014). Therefore our ability to link historical names, and old specimens that are not easily usable, to present collections is limited (De Clerck et al. 2012). Krishnamurthy (1957) provided a complete morphological investigation of field and herbarium material of Bangiopsis including comparisons of the type specimens of B. subsimplex and B. dumontioides (P. L. Crouan & H. M. Crouan) V. Krishnmurthy. He based species distinctness on the cells being slightly larger in B. dumontioides and the cells more widely spaced in B. dumontioides and also the more amount of branching. We do not believe that these characters would be stable to clearly identify these species from the field. Unfortunately no molecular data is available for B. dumontioides. The only non-molecular criteria at the moment to identify these two species is their location in different ocean basins.
West et al. (2005) compared Bangiopsis (now B. franklynottii) from Visakhapatnam, Andra Pradesh, India with Purpureofilum apyrenoidigerum J. A. West, Zuccarello et J. L. Scott from Australia. These two genera seems to be sister species in our phylogenetic analyses. Both genera contain digeneaside and sorbitol as do most genera of the Stylonematophyceae (Karsten et al. 1999, 2003, Zuccarello et al. 2008, Yoon et al. 2010). Purpureofilum apyrenoidigerum chloroplasts lack a pyrenoid whereas those of Bangiopsis have a well-defined central pyrenoid. The ecophysiology of the Indian isolate of B. franklynottii (as B. subsimplex) revealed a wide environmental tolerance characteristic of algae growing in the upper intertidal (Eggert et al. 2007).
Spore formation and germination are very similar in cultured specimens from Australia and India although the mature thalli in the Australian isolate are clearly tubular with a peripheral single-celled layer. The tubular thalli were not evident in the culture of the Indian isolate (West et al. 2005) but Krishnamurthy (1957) observed tubular thalli in field specimens from India.
Bangiopsis subsimplex from the Visakhapatnam coast in India exhibited a diurnal monospore discharge rhythm peaking between 1,300-1,400 h daily and also had maximum discharge at 40 psu salinity (range, 10 to 70) and 50 ℃mol photons m-2 s-1 lighting (range, 0 to 97) (Narasimha Rao and Rangaiah 1991).
As with many of these small red algae their distribution and diversity has been poorly explored. Careful culture observations are needed, along with molecular analysis to confirm species identity (e.g., Zuccarello et al. 2010). Critical evidence on the geographic distribution and genetic variation of Bangiopsis requires more research using these techniques.