The white-spotted flower chafer, Protaetia brevitarsis seulensis, is an insect that belongs to Coleoptera species (Coleoptera: Scarabaeidae). Its life cycle includes the embryo, larva, pupa, and imago stages, of which the larval stage is the longest, where the insect feeds and stores energy for the rest of its life cycle. In Asian countries, P. brevitarsis seulensis larvae are generally known to be pharmacologically effective and used as medicine (Suh et al., 2010). Several reports suggest that P. brevitarsis extract in Lewis’ methanol has antioxidant (Suh et al., 2010), anti-hepatofibrosis, and anti-diabetic properties (Miyanoshita et al., 1996). In addition, scientific reports highlight that, based on the composition of P. brevitarsis seulensis larvae, they can be used as a functional health food. Thus, it is essential to verify that the rearing conditions for P. brevitarsis seulensis larvae are sanitary and safe for consumption as food. On the other hand, Serratia marcescens is a bacterial pathogen, found in soil (Grimont and Grimont, 2006), water (Ajithkumar, 2003; Grimont and Grimont, 2006), and plant material (Grimont and Grimont, 2006). It is also known by alternate names such as Bacillus sphingidis (White, G.F., 1923) and Bacillus asepticus (Burnside, 1928) when it does not show its characteristic red pigment but rather grows as white colonies. S. marcescens and Serratia sp. generally are known to be fatal pathogens of several insect species (Orthoptera, Coleoptera, Hymenoptera, Lepidoptera, and Diptera) once they enter the hemocoel (Tanada et al., 1993). Several reports on Serratia sp. describe severe pathogenic effects on insects (Casper Flyg, 1980; Nunez-Valdez et al., 2008). S. marcescens isolated from Drosophila flies is pathogenic to the insect when ingested with food or when injected into the hemocoel (Casper Flyg, 1980) and is represented as a bacterial pathogen of Rhagoletis pomonella flies (Carol R. Lauzon, 2003). Besides, S. entomophila is pathogenic against the root-damaging larvae of Scarabaeidae (scarab beetles), also called white grubs (Nunez-Valdez et al., 2008). S. entomophila causes Amber disease and is specific to Costelytra zealandica (Hurst et al., 2007a). In addition, S. marcescens can infect mammals and behave as an opportunistic pathogen of man, in addition to plants, insects, and nematodes (Fedrigo et al., 2011). S. marcescens is a gram-negative bacterium belonging to the family Enterobacteriaceae (Gerc et al., 2012). We observed the toxicity of S. marcescens as a fatal insect pathogen and evaluated its lethal dose and concentration when injected into the hemocoel of the 3-stage larvae of P. brevitarsis seulensis. In this work, we have identified the bacterium Spo-1, newly isolated from P. brevitarsis seulensis from Gyeonggi Province, as Serratia marcescens and verified its pathogenicity and toxicity via a bioassay involving intracoelomic injection or oral treatment of P. brevitarsis seulensis larvae with bacterial suspension.
Healthy larvae and those infected with the third larval stage of Protaetia brevitarsis seulensis (Kolbe) were obtained from Gyeong-gi Province, Korea. Larvae were first washed externally with distilled water and 200 μL body fluid was obtained. An aliquot of the body fluid was inoculated in nutrient agar and incubated at 28℃. The single colony, Spo-1 from P. b. seulensis (Kolbe) was isolated and grown in nutrient broth at 28℃.
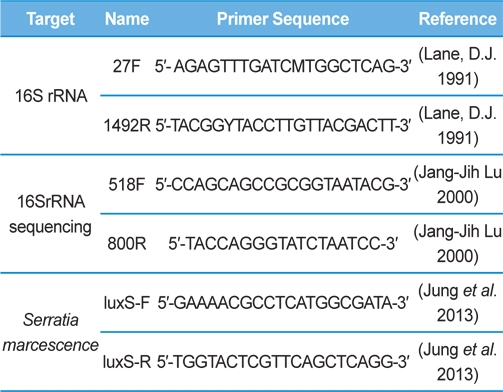
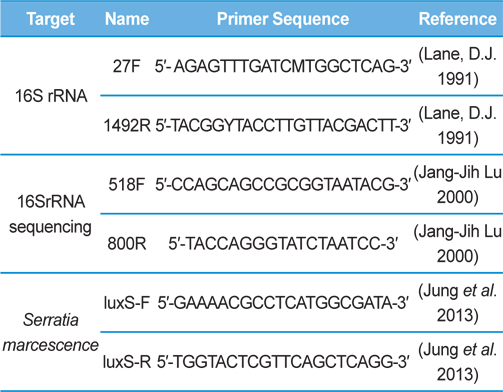
Genomic DNA was extracted from SpoET strain of bacteria from body fluid of P. b. seulensis (Kolbe) using a Wizard PCR Preps DNA Purification System (Promega, Madison, WI, USA). The 16S small subunit ribosomal RNA (16SrRNA) gene was amplified by PCR using universal primers 27F (5′- AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′). The PCR products were purified on 1% agarose gel and sequenced using universal primers for 16SrRNA sequencing, 518F (5′-CCAGCAGCCGCGGTAATACG-3′) and 800R (5′-TACCAGGGTATCTAATCC-3′) designed by Macrogen co. (Seoul, Korea). Sequence alignment was compared using Basic Local Alignment Search Tool (BLAST) of National Center for Biotechnology Information (NCBI, USA) and ClustalW (Multiple Sequence Alignment, UK) .
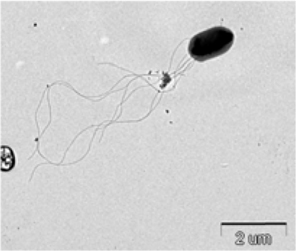
The bacterium isolated from P. b. seulensis (Kolbe) was named as Spo-1. The third-stage P. b. seulensis (Kolbe) larvae were dissected in distilled water in order to observe the Spo-1 structure by electron microscopy. Sections for electron microscopy were mounted onto 200-mesh Ni grid, and viewed using H-7000 Hitachi microscope (Hitachi, Japan) at 10 kV and 7.5 kV.
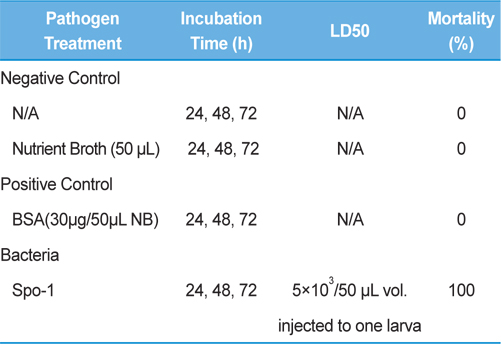
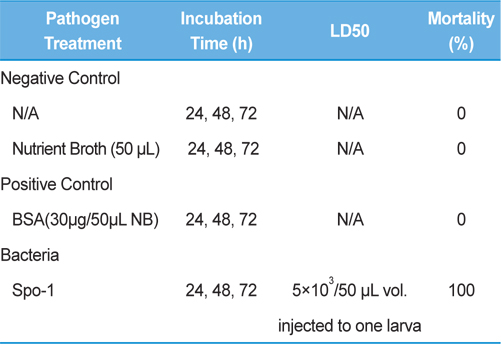
Bioassay was performed by intracoelomic injection of 50 μL bacterial suspension (101 to 107 bacteria per larva) in nutrient broth. Hemocytometer was used for cell counts. Negative control larvae were injected with the same volume of sterile nutrient broth and positive control using bovine serum albumin (for concentration standard and same proportionate with bacteria concentration) larvae were injected with the same volume of bovine serum albumin (BSA) (30 μg/50 μL per larva) into the hemocoel. Mortality was recorded at 24, 48, 72-h intervals. Injected larvae were reared at 24℃. The oral treatments doses comprised of 1011/50 mL volume of bacteria mixed with fermented sawdust as food to 10 larvae in rearing box. Bioassay tests with oral or injection treatment were performed in triplicate, using 7 to 10 healthy, second- or third-stage larvae per test.
Genomic DNA from larvae infected with Spo-1 from P. b. seulensis (Kolbe) was extracted and suspended in 500 μL double-distilled water and centrifuged at 3000 rpm (100.8g) for 5 min. The pellet was used to purify genomic DNA using Wizard PCR Preps DNA Purification System (Promega, USA). PCR reaction was carried out in a 25-μL volume using PCR Premix kit (Bioneer, Korea). To extract 16SrRNA from Spo-1, DNA amplification was performed as follows: initial denaturation at 94℃ for 3 min, 35 cycles of denaturation (1 min at 94℃), annealing (1 min at 57℃), extension (1 min at 72℃), followed by final extension at 72℃ for 10 min. Seven samples from each amplified PCR run was separated on 1% agarose gel with ethidium bromide staining, in 1xTEA buffer and visualized using a UV transluminator. PCR products were sequenced using Macrogen co. (Seoul, Korea) and suggested nucleotide sequences were identified by Basic Local Alignment Search Tool (BLAST). The 16SrRNA gene was amplified using the primers: luxS-F (5′-GAAAACGCCTCATGGCGATA-3′), luxS-R (5′-TGGTACTCGTTCAGCTCAGG-3′) (Jung et al., 2013, Joyner et al., 2014) (Table 1). PCR was performed as follows: initial denaturation at 94℃ for 5 min, 30 cycles of denaturation (30 s at 94℃), annealing (30 s at 56℃), extension (30 s at 72℃), followed by the final extension at 72℃ for 5 min (Jung et al., 2013).
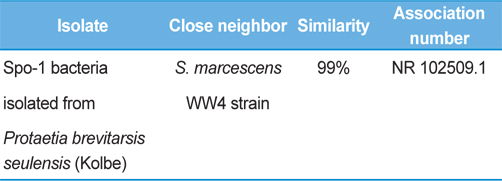
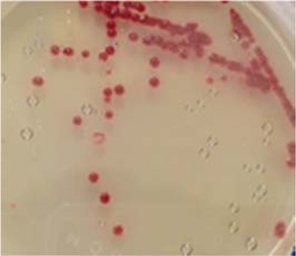
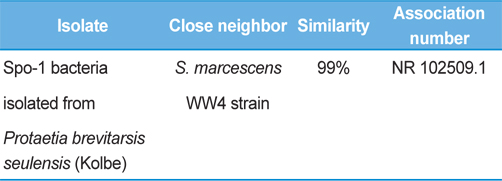
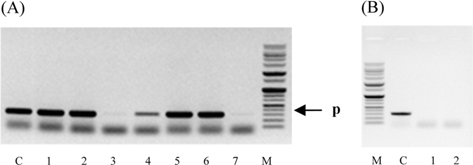
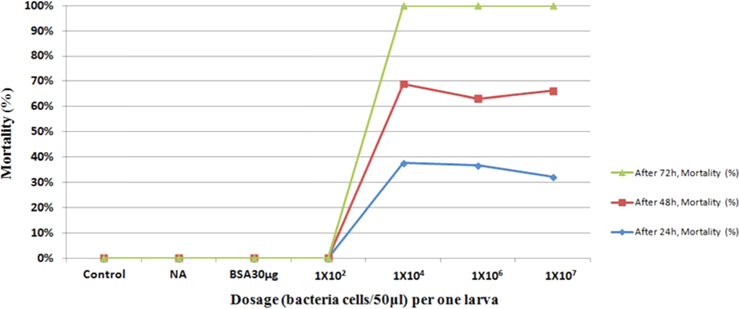
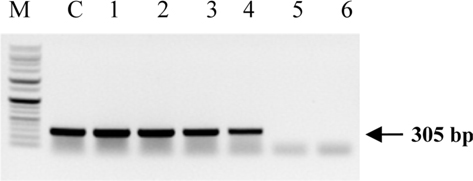
Symptoms of bacteria-infected Protaetia brevitarsis seulensis (Kolbe) larvae included black spots of varying size appearing on the body. Small-sized (1 to 3 mm × 1 to 3 mm) black spots on the whole body or at one spot (5 mm to 1 cm × 5 mm to 1 cm) located dorsally, at the base of the P. b. seulensis larval body. Diseased larvae generally showed transparent black-brown color and moved insensitively and inactively and reacted slowly to touch. After observing the diseased larvae, we isolated bacterial cells from them, incubated on nutrient agar plates at 26℃ and named the bacterium as Spo-1. Cells of strain Spo-1T were rod shaped and colonies showed bright-red pigment on nutrient agar plate after 24 h. Transmission electron microscope (TEM) images of Spo-1 showed rod-shaped body and several long flagella, 1.5 to 2 × 6 to 7 μm in size, similar to Serratia marcescens (Fig. 3). To identify Spo-1 species, 16SrRNA sequences are analyzed. Primer sets used are shown in Table 1. And genomic DNA of bacteria was amplified almost the full length of the 16SrRNA, 1492-1500 bp. PCR amplification was performed in a total volume of 20 μL containing 50 ng bacterial genomic DNA extract and 10 pmol of each primer. The 16SrRNA sequences were highly conserved and PCR products contained approximately 1500 bp. The 16SrDNA sequence of isolate Spo-1 showed 99% similarity to S. marcescens, NCBI accession number NR 102509.1. (Table 2). The 7 diseased P. b. seulensis larvae in each samples were washed with distilled water several times and the mucus membrane lining the hemocoel from the larvae was collected. We extracted genomic DNA from the hemocoel of diseased larvae and PCR was performed to detect 16SrRNA bands using universal primers; 27F and 1492R (Table 1). After PCR amplification, we found that S. marcescens was not detected in the hemocoel of normal P. b. seulensis larvae. However, the diseased larvae samples showed the presence of S. marcescens, identified via the 305-bp bands. A native bacterial strain, Spo-1 was isolated from the hemocoel of infected third-stage larvae of P. b. seulensis that appeared to have amber disease. The larvae were collected from Gyeong-gi Province, Korea. Intracolelomic injection assay was done to evaluate the fatal effects of Spo-1 on the P. b. seulensis larvae and document pathogenic symptoms. Normal larvae of P. b. seulensis were injected with different concentrations of bacteria (from 1 × 102/50 μL to 1 × 107/50 μL) in nutrient broth using Bovine serum albumin as control (30 μg/50 μL). The larvae injected with bacteria were observed during 24, 48, and 72 h after intracoleomic injection and symptoms showed significant similarity to diseased larvae of P. b. seulensis. The concentration of bacterial cells (1 × 104/50 μL) exerted significant effect and showed 100% mortality in the third-stage larvae of P. b. seulensis after 72 h (Fig. 3). In addition, when 1 × 107 cells of Spo-1 were injected into the larva, mortality of P. b. seulensis larvae was 100% after 24 h. Additionally, the LD50 of Spo-1 was 5 × 103 cells/ μL (Table 3). To evaluate the specificity and sensitivity of PCR product of Spo-1, we tested the band sizes according to the concentration of Spo-1. The tested concentrations were 50, 25, 5, 0.5, 0.05, and 0.005 ng/μL. We could detect the band until 0.5 ng/μL Additionally, the concentration of the bacterial samples correlated with the band size.
In Korea, the white-spotted flower chafer (P. b. seulensis) larva is known to have pharmacological activity in diseases such as hepatic disorders (Kwon et al., 2013). This insect is being reared artificially to meet the demand for this novel, functional food in Korea. This mass conditional rearing might expose the larvae to contamination due to inadequate sterilization, disinfection, or other systematic rearing conditions. Therefore, it is essential to set up sanitary rearing conditions to supply healthy insects as food. Sawdust, used as feed for larvae of the white-spotted chafer, is rough, so the epidermis of the larvae may get scratched and infected by many kinds of bacteria, including fatal insect pathogens. In this study, we report S. marcescens infection in the third-stage P. b. seulensis larvae and S. marcescens is found to be fatal to the larvae. This bacterium can be pathogenic to humans through the oral route (Carol R. Lauzon, 2003; Schwarz, 2004). S. marcescens is commonly found in soil, water, plant material, and mammals (Joyner et al., 2014). The common bacterial pathogens of insects include S. marcescens (Ajithkumar, 2003; Carol R. Lauzon, 2003; Lauri Mikonranta, 2014; Murdoch et al., 2011), S. entomophila (Hurst et al., 2007a; Hurst et al., 2007b; Nunez-Valdez et al., 2008), Bacillus thuringiensis (Arthur I. Aronson, 1986; El-Aasar A.M.; El-Sheikh, 2013), and Pseudomonas aeruginosa (Martinez-Granero et al., 2014; Mulcahy et al., 2011). In this study, we isolated a bacterial strain from the white-spotted flower chafer in Gyeong-gi Province, Korea, and named it Spo-1. Spo-1 was cultured in nutrient agar plates, and after incubation, it showed typical morphology when visualized under the electron microscope. We observed that Spo-1 have many flagella at the end of its oval-shaped body (Fig. 3). In addition, by using 16srRNA PCR and sequencing, we confirmed that the larvae of P. b. seulensis from a rearing laboratory in Gyeong-gi are infected with Spo-1(S. marcescens) (Table 2). For more specific identification of Spo-1, we assayed PCR with specific primers and identified that Spo-1 is certainly S. marcescens. In addition, bioassay of Spo-1(intracoleomic injection to larvae) revealed that Spo-1 have the mortality when it treated to third stage of larvae and showed the percentage of LD50 (lethal dose) is 5 × 103/50 μL per larva and LT50 (lethal time) is 18 h when injected with 1 × 104/50 μL per larva (Fig. 4), (Table 3). After injecting Spo-1 into the larvae, we re-identified that Spo-1 from larva’s hemocoel and the bacteria are Spo-1. Furthermore, to evaluate the possible concentration of Spo-1, we detected PCR bands from 0.5 ng/μL genomic DNA of the infected larvae hemocoel (Fig 7). We highlight the importance of sanitary rearing conditions for P. b. seulensis larvae in order to produce healthier and effective functional food. Therefore, it is essential to establish systematic and sanitary rearing environments for these insects to avoid deleterious effects on the health of consumers after consumption of infected larvae. These findings suggest that unfavorable rearing conditions, including rough sawdust, less ventilation, and lessly exchange sawdust periodically as feeding may causes diseases in the insect larvae, and also the epidermis of larvae may get scratched by the rough sawdust, thereby causing bacterial infection.